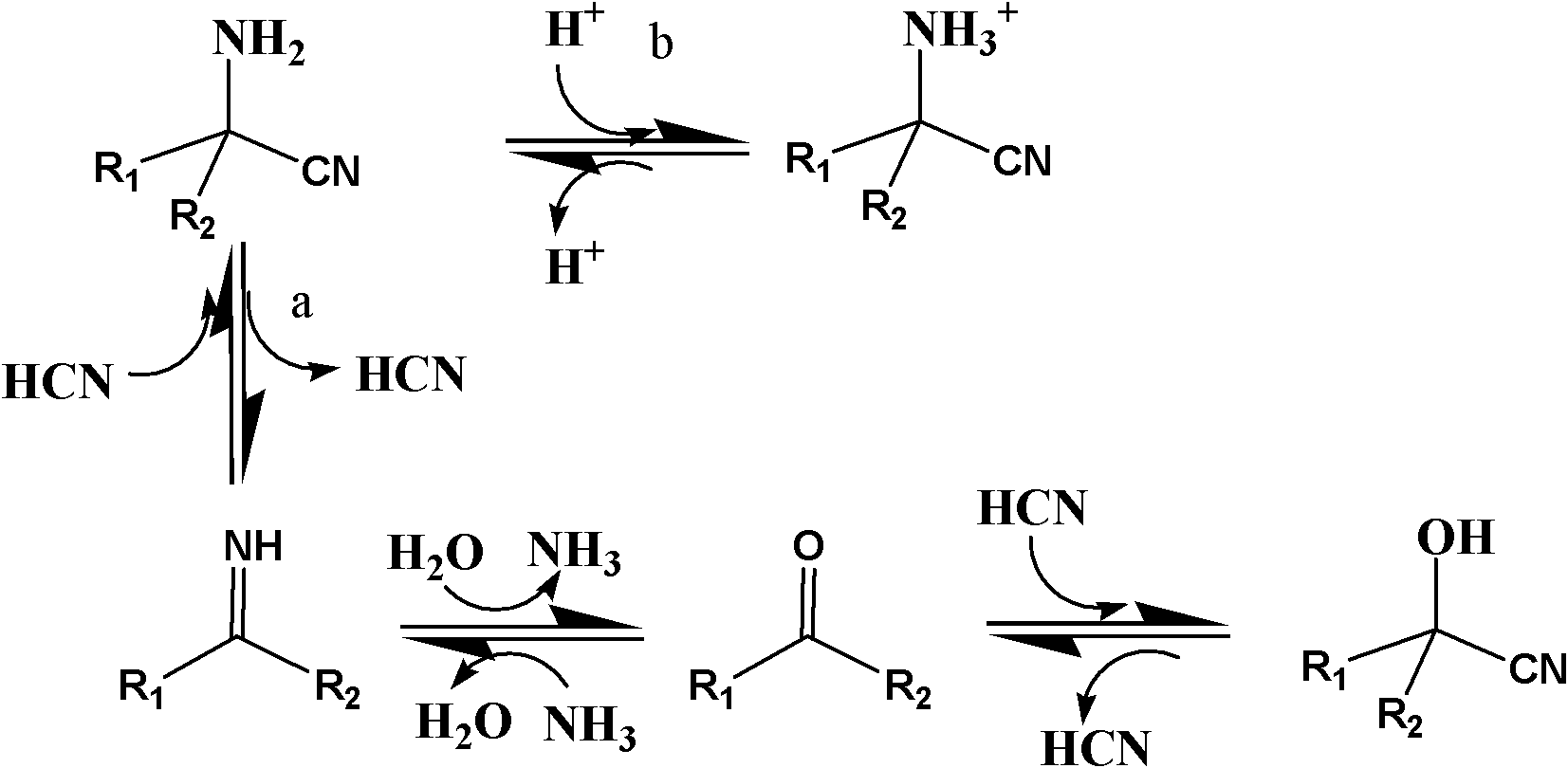
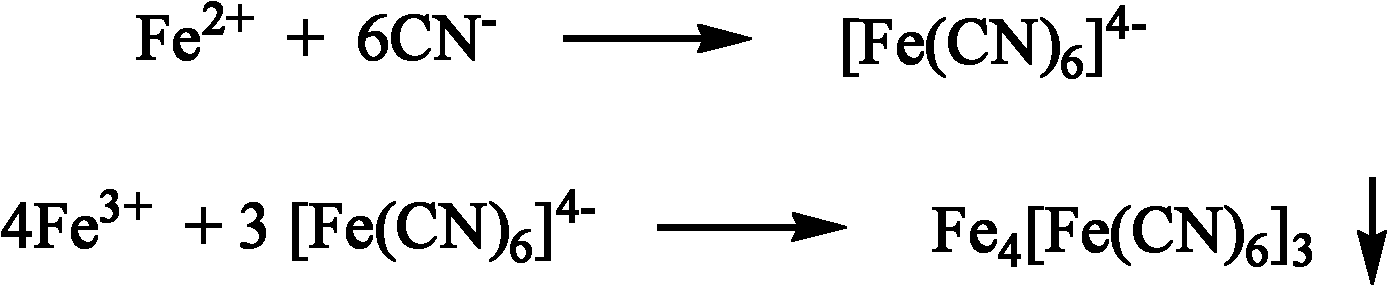
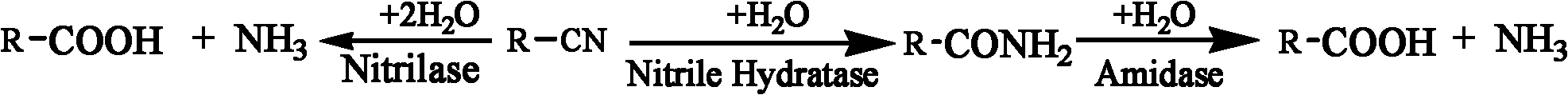High-throughput screening method of nitrile invertase
A technology for nitrile converting enzymes and screening methods, which is applied in the field of high-throughput screening of nitrile converting enzymes, can solve the problems of poor versatility, high equipment requirements, time-consuming and laborious, etc., and achieve the effects of simple operation, rapid detection, and convenient rapid screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Screening of nitrile hydratase-producing bacteria containing 2-amino-2,3-dimethylbutyronitrile hydration activity
[0038] After culturing the microorganisms to be screened, they were centrifuged, washed with normal saline, evenly suspended in distilled water, and prepared as OD 600 =10 bacterial suspension. Take 5ml of the bacterial suspension and place it in a stoppered Erlenmeyer flask (50ml), then put it in a 30°C water bath to preheat for 5min, and add 20μl of 2-amino-2,3-dimethylbutyronitrile to start the reaction. Samples were taken after 15 minutes, and centrifuged at 10,000 rpm for 5 minutes. Take 100 μl of the supernatant in a 96-well plate, and add FeSO with a concentration of 100 mM successively 4 and FeCl 3 100 μl of each aqueous solution, and observe the color change. Joined FeSO successively 4 and FeCl 3 After aqueous solution, the experimental group whose supernatant first changed from colorless to light green and then to yellow containe...
Embodiment 2
[0040] Example 2: Screening of nitrile hydratase-producing bacteria containing β-aminopropionitrile hydration activity
[0041] After culturing the microorganisms to be screened, make a bacterial suspension according to the method of Example 1, take 10ml of the bacterial suspension and place it in a stoppered Erlenmeyer flask (50ml), then put it in a water bath at 20°C for preheating for 10min, add 30μl of β -Aminopropionitrile starts to react. Samples were taken after 10 minutes, and centrifuged at 12,000 rpm for 4 minutes. Take 200 μl of the supernatant in a 96-well plate, and add FeCl with a concentration of 1.0M successively 2 and Fe(NO 3 ) 3 20 μl of each aqueous solution, and observe the color change. Add FeCl successively 2 and Fe(NO 3 ) 3 After aqueous solution, the experimental group whose supernatant first changed from colorless to light green and then to yellow contained nitrile hydratase.
[0042] Controlled by colorimetry, the strains Rhodococcus sp.ZJB-09...
Embodiment 3
[0043] Example 3: Screening of amidase-producing bacteria containing 2-aminobutyramide hydrolysis activity
[0044] After culturing the microorganisms to be screened, make a bacterial suspension according to the method of Example 1, take 7ml of the bacterial suspension and place it in a stoppered Erlenmeyer flask (50ml), then put it into a water bath at 40°C for preheating for 4min, add 20 μl of 2 -Aminobutanamide starts to react. Samples were taken after 40 min, and centrifuged at 10,000 rpm for 6 min. Take 180 μl of the supernatant in a 96-well plate, and add FeCl at a concentration of 200 mM 2 and Fe(NO 3 ) 3 60 μl of each aqueous solution, and observe the color change. Add FeCl successively 2 and Fe(NO 3 ) 3 After aqueous solution, the supernatant does not undergo a significant color change and eventually shows Fe 3+ The yellow test group of the ionic solution contains amidase.
[0045] The strain Delftia tsuruhatensis ZJB-05174 (CCTCC No: M 205114) can catalyze t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com