Method for rapidly determining drug tolerance of strain
A rapid determination and drug resistance technology, applied in the field of rapid determination of bacterial resistance, rapid determination of bacterial susceptibility to antibiotics, and can solve problems such as inability to use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0024] According to one embodiment of the rapid determination method of bacterial strain drug resistance of the present invention, the determination method comprises the steps:
[0025] A. Prepare thiazolyl blue with PBS to make a 5±0.5% stock solution, sterilize it, and store it in a refrigerator at about 4°C. Add the thiazole blue stock solution to the brain heart infusion culture solution to make the final concentration reach 0.1±0.05%, sterilize it, and store it in a refrigerator at about 4°C for later use;
[0026] B. Prepare a bacterial suspension of 1-10CFU / ml;
[0027] C. prepare antibiotic solution;
[0028] D. Add 178 ± 20 μl of brain heart infusion containing thiazolium blue to each well of the culture plate, and add 20 ± 5 μl of bacterial suspension, and then add 2 ± 1 μl of antibiotics, serially dilute the antibiotic solution, and the final concentration of the antibiotic solution is a series of concentrations, such as 0.1, 1, 10 and 100 μg / ml series concentrati...
Embodiment 1
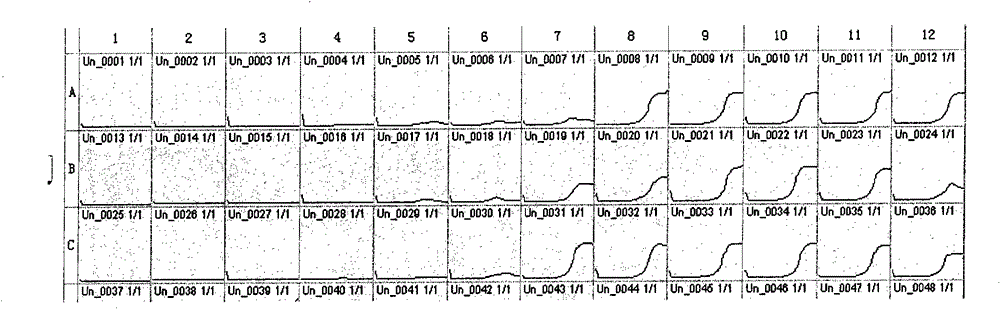
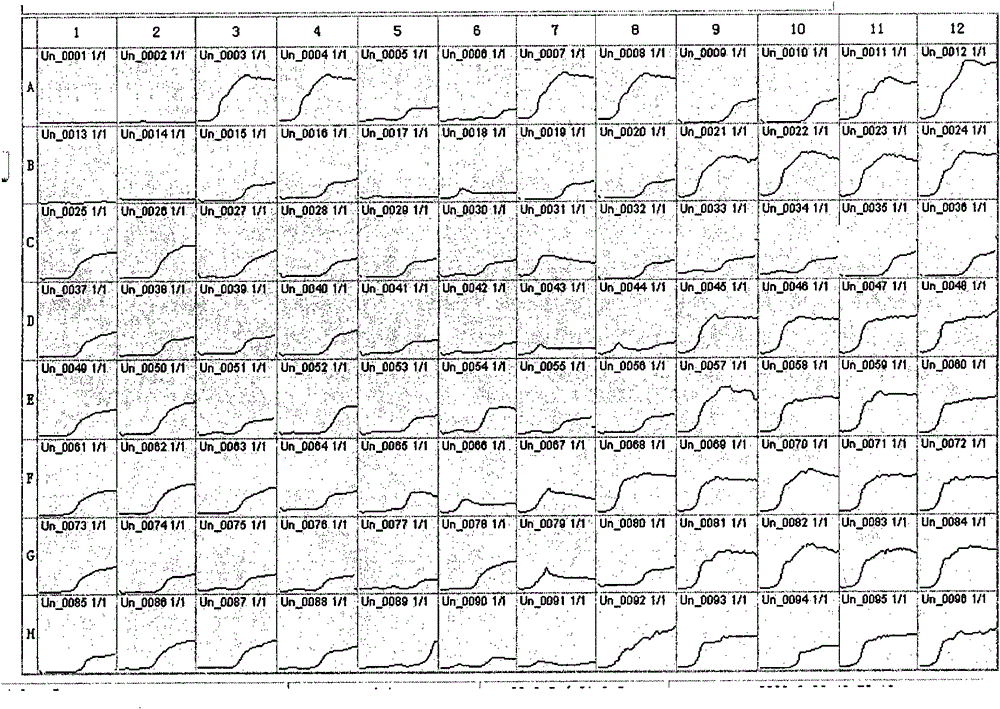
[0034] Take Staphylococcus aureus as an example of a Gram-positive bacterium. The thiazolyl blue was prepared into a 5% stock solution with PBS, sterilized, and stored in a refrigerator at 4°C. The thiazole blue stock solution was added to the brain heart infusion culture solution to make the final concentration reach 0.1%, sterilized, and stored in a 4°C refrigerator for later use. Add 178 μl of brain heart infusion containing thiazolium blue to each well of a 96-well culture plate. Staphylococcus aureus (strain IQCC22004) bacterial suspension was prepared so that its concentration reached the order of 10 CFU / ml, and 20 μl of bacterial suspension was added to the microwells. Among them, the three lines of A, B, and C were repeated three times, A1, B1, and C1 were blank controls, and 2 μl of sterile water was added; A2, B2, and C2 were added with 2 μl of amoxicillin solution with a concentration of 100 mg / ml, so that The final concentration is 1mg / ml; A3, B3, C3~A11, B11, C1...
Embodiment 2
[0037] Take Enterobacter sakazakii as an example of a Gram-positive bacterium. Add 178 μl of brain heart infusion containing thiazolium blue to each well of a 96-well culture plate. Prepare a bacterial suspension of Enterobacter sakazakii (strain IQCC 10424) so that its concentration reaches the order of 10 CFU / ml, and add 20 μl of the bacterial suspension to the microwells. Repeat the test with two wells. A1 and A2 are negative controls, add 20 μl of sterile water instead of bacterial suspension; A3 and A4 are blank controls, add 2 μl of sterile water instead of antibiotics. A5, A6, A7, A8 are 0.1 μg / ml, 0.05 μg / ml vancomycin, and so on, A9, A10~H11, H12 are oxacillin, cefazolin, cefpodoxime, cephalothin, penicillin G , cefoxitin, amoxicillin, ampicillin, imipenem, pefloxacin, gentamicin, carbenicillin, netilmicin, cefotaxime, cefnicillin, nitrofurantoin, compound Sumophen, ciprofloxacin, ceftazidime, norfloxacin, tetracycline, tobramycin solution. All the other steps a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com