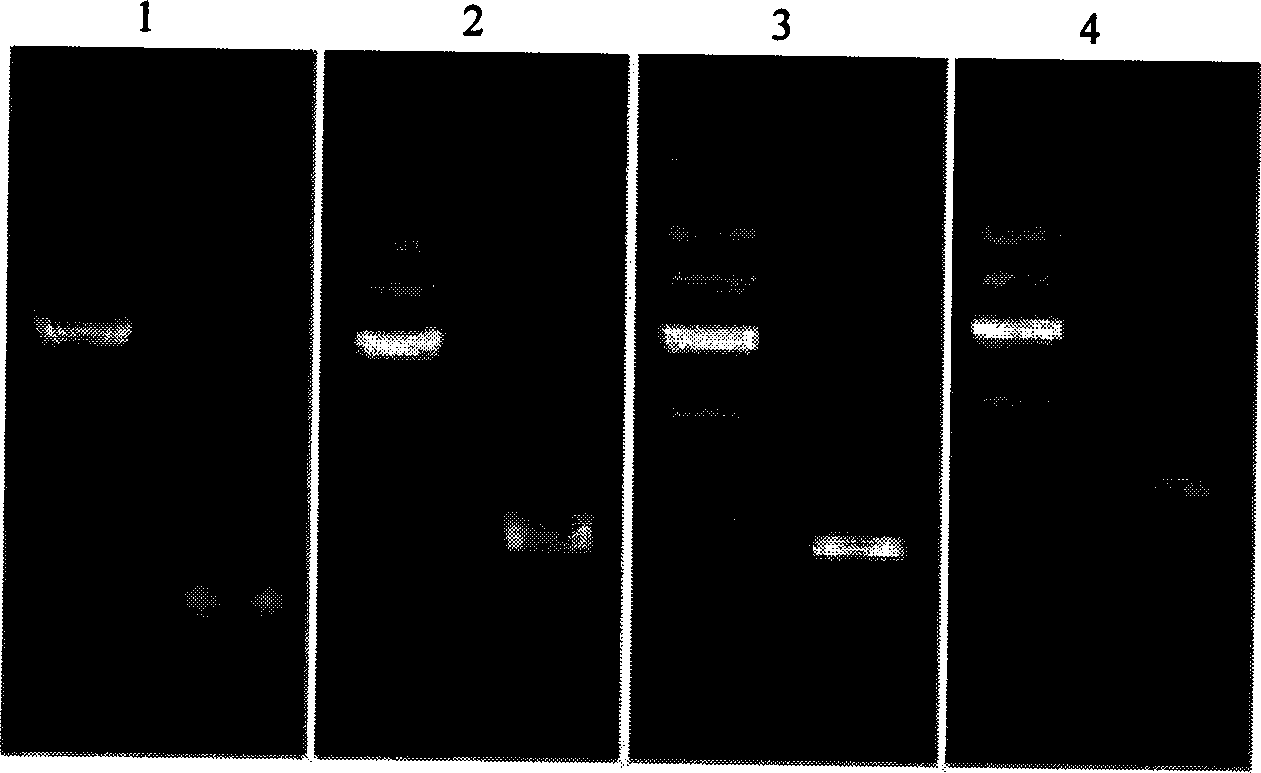
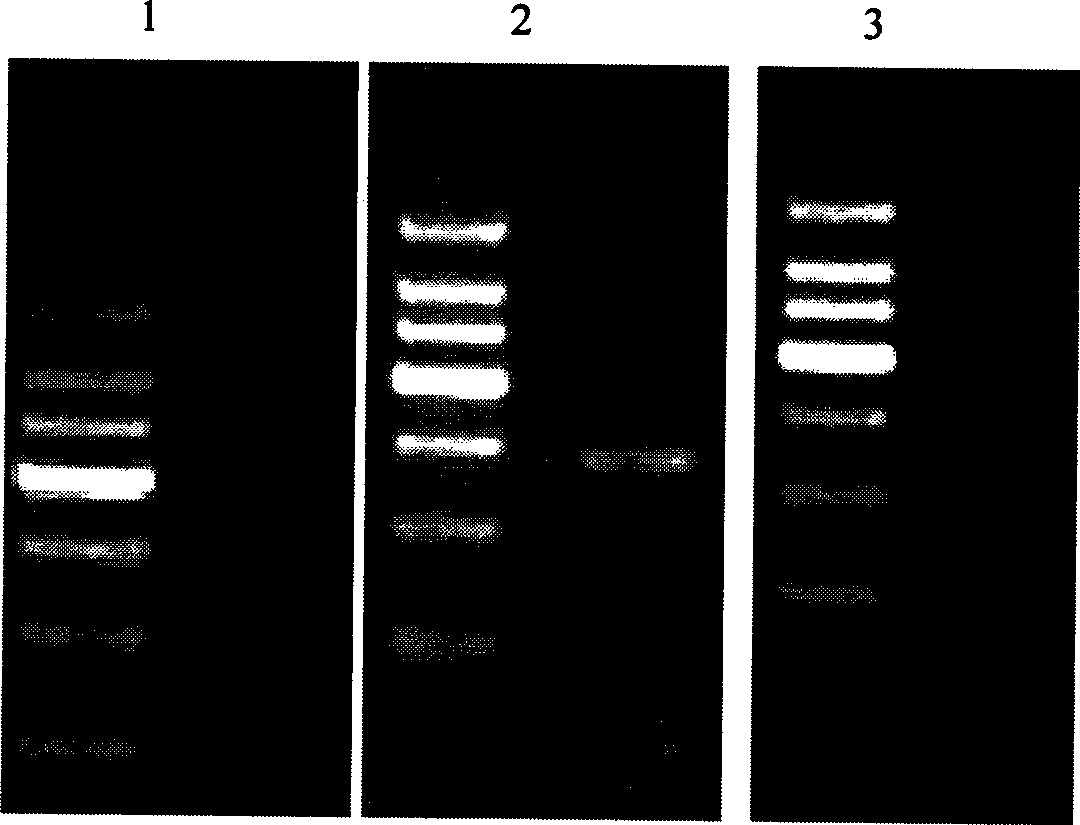
DNA immobilization method based on nucleic acid immobilization by gel and use thereof
An immobilization method and nucleic acid technology, applied in the field of target nucleic acid detection, can solve problems such as the reduction of immobilization efficiency, and achieve the effects of eliminating possibility, high immobilization efficiency and high loading capacity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Detection of Efficiency of DNA Immobilization by Hydrogel Copolymerization
[0040] (1) Preparation of end-modified PCR amplification products
[0041] Using the plasmid DNA containing the Staphylococcus aureus enterotoxin E gene (pUCm-T-SEE, see GenBank accession number M21319) as a template, a fragment of about 850 bp of the gene was amplified by PCR. The base sequences of the two primers used for this PCR are shown in SEQ ID NO.1 and SEQ ID NO.2; during primer synthesis, (CH2) was introduced at the 5' end of the upstream and downstream primers 6 -NH 2Modification; after the PCR reaction is completed, the PCR product is recovered, quantitatively analyzed by GeneQuant, and stored at -20°C for later use;
[0042] (2) Take 5% polymethacrylamide gel and 20% polymethacrylamide gel for use; configure the gel according to the following formula
[0043] 5% polymethacrylamide gel 20% polymethacrylamide gel
[0044] h 2 O 3.167mL —
[0045] 10×TBE(pH1...
Embodiment 2
[0054] Thermal Stability Experiment of Fixation Method
[0055] (1) Preparation of end-modified PCR amplification products
[0056] Using the plasmid DNA (pUCm-T-SEE, GenBank accession number M21319 of which see) containing the Staphylococcus aureus enterotoxin E gene as a template, a fragment of about 850 bp of the gene was amplified by PCR; two primers for this PCR See SEQ ID NO.1 and SEQ ID NO.2 for the base sequence; in the primer synthesis process, (CH2) was introduced at the 5' end of the upstream and downstream primers 6 -NH 2 Modification; after the PCR reaction is completed, the PCR product is recovered, quantitatively analyzed by GeneQuant, and stored at -20°C for later use;
[0057] (2) Immobilization of PCR products
[0058] Take the previously amplified and purified PCR product (with NH at the end 2 Modification), take 1 μL and dilute to 10ng-1μg / μL with double-distilled water; take the diluted PCR product, heat and denature it at 100°C for 5 minutes, and imme...
Embodiment 3
[0070] Immobilization of pUCm-T-SEE plasmid fragment
[0071] (1) Fragmentation and terminal modification of pUCm-T-SEE
[0072] Extract the pUCm-T-SEE plasmid according to the conventional method, take 100 μL of the purified pUCm-T-SEE plasmid, add 50 μL of formic acid, mix well and place it in a water bath at 30°C for a reaction time of 20 minutes; add NaOH milk immediately after the reaction Adjust the pH of the turbid solution to 7.0; according to the kit instructions, use the DNA gel recovery kit to recover the plasmid DNA after formic acid treatment, recover the DNA on the column with 100 μL of double distilled water, and the liquid in the centrifuge tube is the recovered plasmid DNA ; Add 25 μL of 2.5mol.L to the depurinated plasmid solution -1 ethylenediamine hydrochloride solution, after mixing, add NaOH emulsion to adjust the pH to 7.4, and react in a water bath at 37°C for 4 hours; after 4 hours, add 25 μL of freshly prepared 0.1mol L -1 Sodium borohydride solutio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com