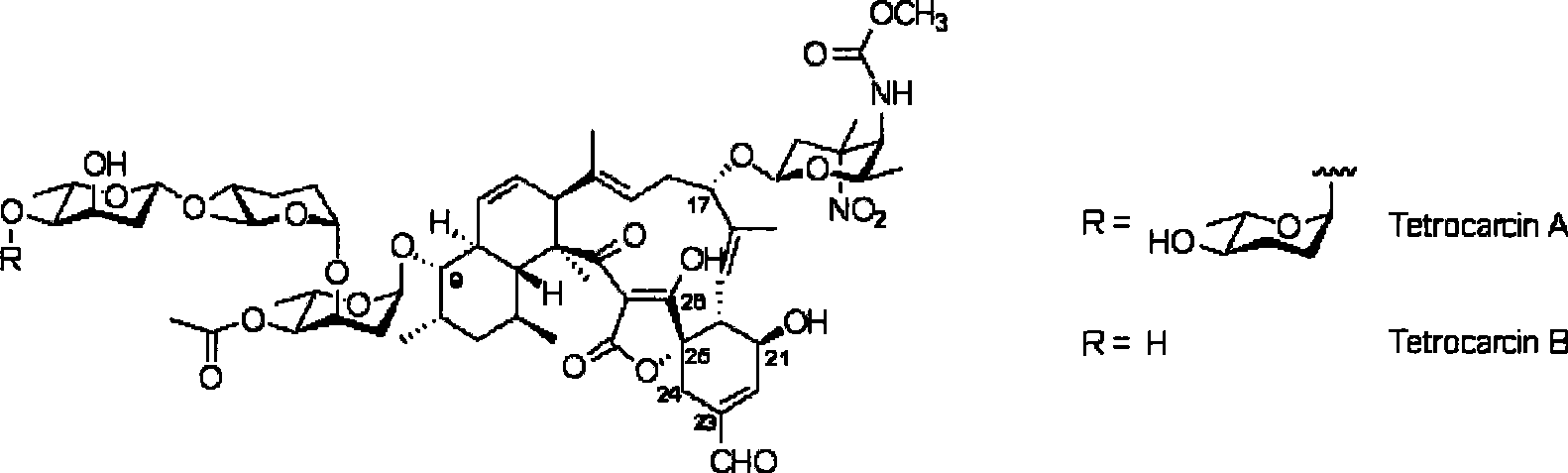
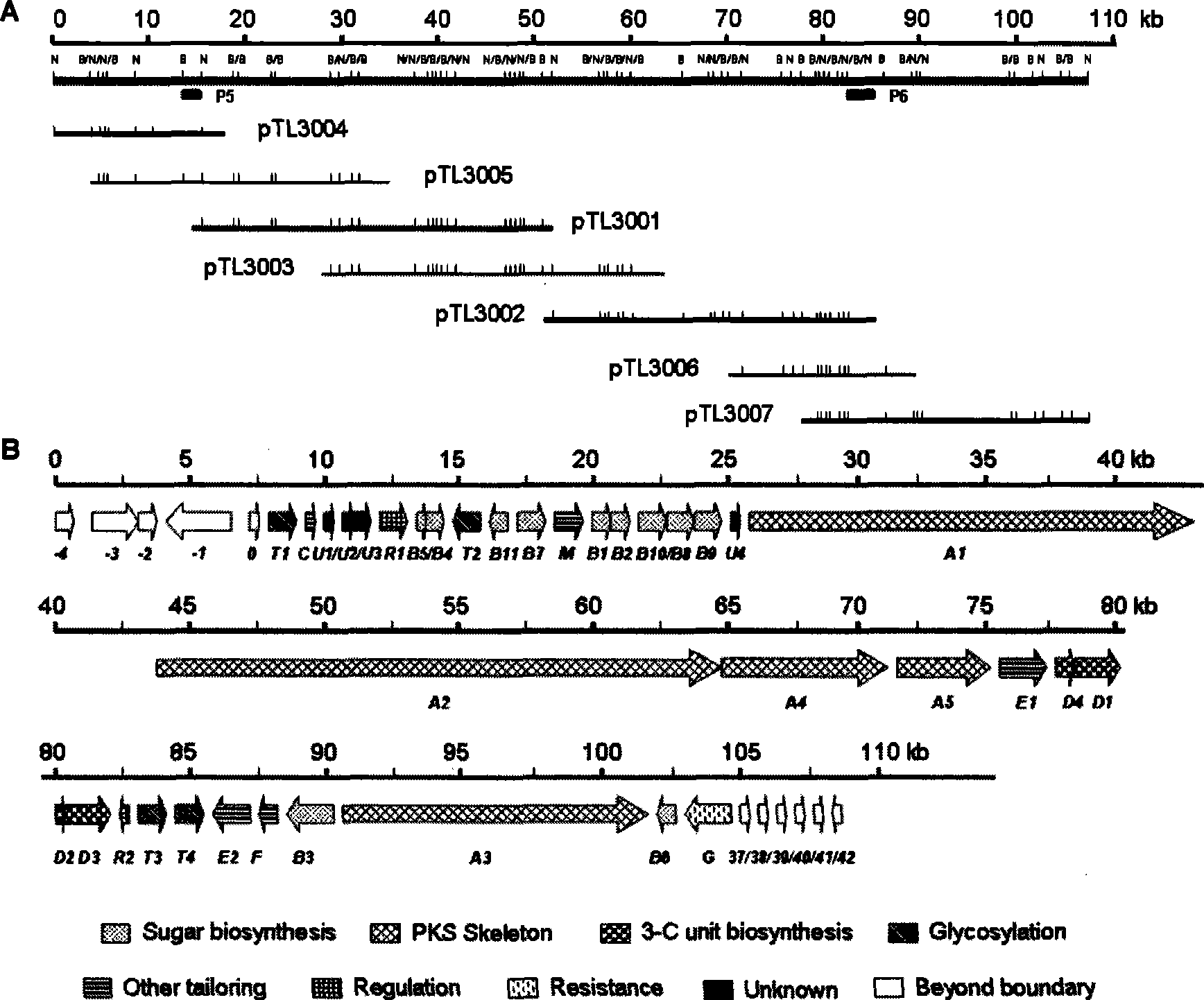
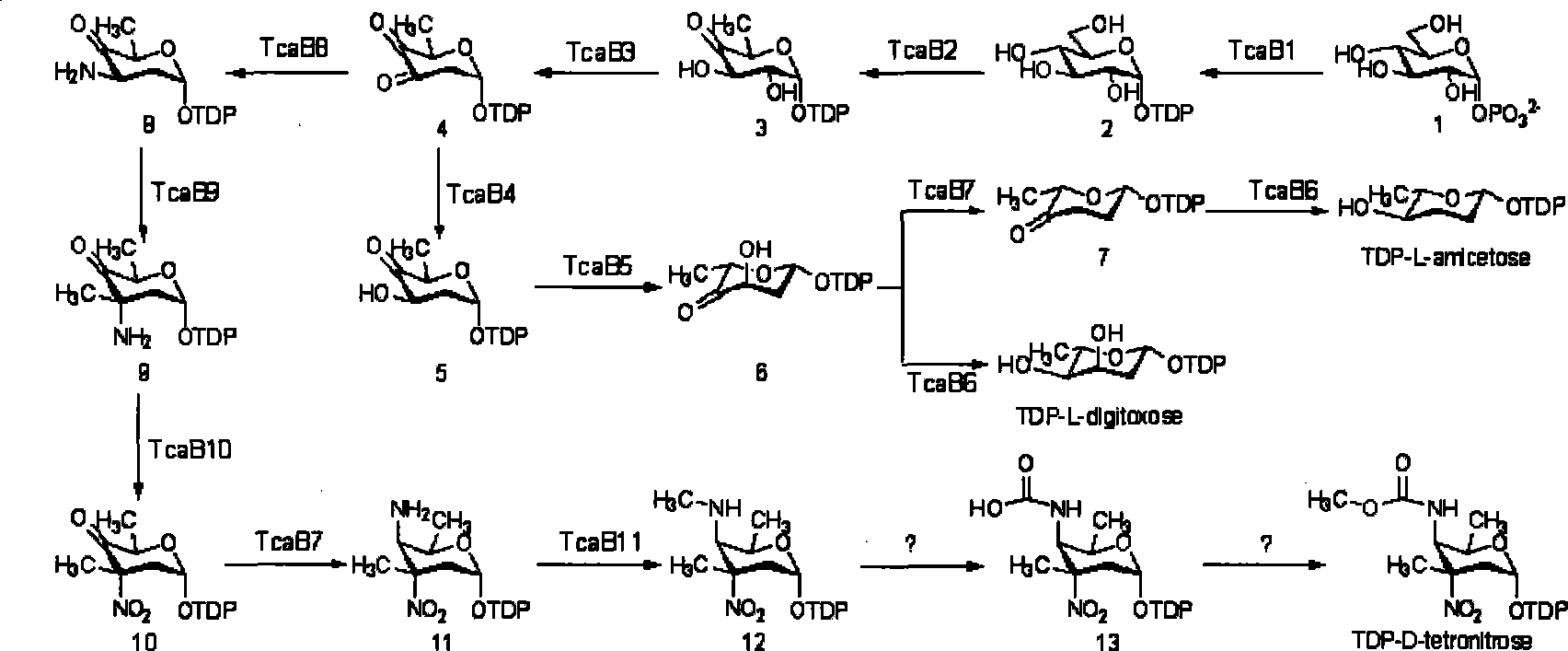
Biological synthesis gene cluster of tetrokacin A and use thereof
A technology for biosynthesis and tetrikacin, which is applied in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc., and can solve the problems of long chemical semi-synthetic route and low yield.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Extraction of total DNA of Tetrocacin A-producing bacteria Micromonospora cyanococcus NRRL11289:
[0097] Draw 100 μl of -80℃ frozen Micromonospora mycelium suspension and inoculate it into a medium containing 3ml TSB (30g / L T krypton S oya B roth) culture medium, 30°C, shaking culture for about 36 hours, take 2.5ml of bacterial liquid and inoculate into 50ml TSB (containing 0.5% glycine, 25mM magnesium chloride) medium, 30°C, shaking culture for about 36 hours until the bacteria The liquid was fresh orange, and the cells were collected by centrifugation at 3500 rpm, washed twice with lysate (150 g / L sucrose, 25 mM Tris-Cl, 25 mM EDTA), and stored at -20°C for extraction of genomic DNA. Add 10ml of lysate (containing lysozyme 5mg / ml, achromogenic peptidase 1mg / ml) to the collected hyphae, vortex until uniform, incubate at 37°C for 30 minutes, add 0.1ml proteinase K (4mg / ml), 1ml 10% SDS, mix well and quickly place in a 70°C water bath for 15 minutes until the solutio...
Embodiment 2
[0099] Construction of Genome Cosmid Library of Tetrocacin A-producing Micromonospora cyanococcus NRRL11289:
[0100] First, through a series of dilution experiments to determine the amount of Sau3AI, on this basis, a large number of enzyme-digested DNA fragments slightly larger than 40kb, dephosphorylated. pOJ446 was first cut and dephosphorylated from the middle of the two cos sequences with BglII, and then cut with BamHI from the multiple cloning site to obtain two arms, which were ligated with the prepared 40kb DNA fragment overnight. Take out the packaging mixture from the -80°C refrigerator and place it in a dry ice bucket. Melt the packaging mixture quickly between your fingers. Add 4 μL of the ligation product when it just starts to melt, mix well with a gun, and place in a 22°C water bath for 2 hours. Add 500 μL SM buffer [(L -1 ): 5.8g NaCl, 2.0g MgSO 4 , 50.0ml of 1M Tris.HCl (pH=7.5), 5.0ml of 2% (w / v) agar] and 50 μL of chloroform, mixed gently, and centrifuged ...
Embodiment 3
[0105] Nucleic acid molecular hybridization:
[0106] 1) DIG DNA labeling: Dilute the DNA to be labeled with sterile water to a total volume of 15 μL, heat and denature it in a boiling water bath for 10 minutes, and immediately place it in an ice-salt bath to cool. Then add 2 μL of primer mixture, 2 μL of dNTP mixture, and 1 μL of enzyme, mix well, and place in a 37° C. water bath for about 16 hours. Add 0.8 μL 0.8M EDTA (pH 8.0) to terminate the reaction, add 2.5 μL 4M LiCl and mix well, then add 75 μL pre-cooled absolute ethanol to precipitate the labeled DNA, and place it at -80°C for 40 minutes. The DNA was collected by centrifugation at 12000 rpm for 20 minutes at 4°C, washed with pre-cooled 70% ethanol, dried in vacuo and redissolved in 50 μLTE (pH 8.0).
[0107] 2) Quality detection after DIG DNA probe labeling: Dilute the labeled DNA probe to the following six gradients, 1, 10 -1 、10 -2 、10 -3 、10 -4 、10 -5 . Dilute the labeled control DNA to the following conce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com