Recombined endo-chitinase gene order and recombinant vector threreof
A technology of chitinase gene and recombinant vector, which is applied in the field of genetic engineering, can solve the problems of difficulty in controlling the degree of degradation, high energy consumption of acid degradation, secondary pollution of the environment, etc., and achieve efficient and cheap industrial production, easy purification, The effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0010] Example 1. Acquisition of recombinant endochitinase gene sequence based on Pichia pastoris codon bias
[0011] According to the codon preference of Pichia pastoris and the gene sequence of endochitinase, the following primers were designed:
[0012] f 1 : 5'-GAATTCGCTAGTGGTTACGCTAACGCTGTTTACTTTACTAACTGGGGTATTTACGGT-3'
[0013] f 2 : 5'-ACTGGGGTATTTACGGTCGTAACTTTCAACCACAAAACCTTGTTGCTTCTGATATTACT-3'
[0014] f 3 : 5'-TGTTGCTTCTGATATTACTCATGTTATTTACTCTTTTATGAACTTTCAAGCTGATGGTACT-3'
[0015] f 4 : 5'-TTCAAGCTGATGGTACTGTTGTTTCTGGTGATGCTTACGCTGATTACCAAAAGCATTAC-3'
[0016] f 5 : 5'-ATTACCAAAAGCATTACGATGATGATTCTTGGAACGATGTTGGTAACAACGCTTACGGT-3'
[0017] f 6 : 5'-GTAACAACGCTTACGGTTGTGTTAAGCAACTTTTTTAAGTTGAAGAAGGCTAACCGTAAC-3'
[0018] f 7 : 5'-AGAAGGCTAACCGTAACTTGAAGGTTATGCTTTCTATTGGTGGTTGGACTTGGTCTACT-3'
[0019] f 8 : 5'-GTTGGACTTGGTCTACTAACTTTCCATCTGCTGCTAGTACTGATGCTAACCGTAAGAAC-3'
[0020] f 9 : 5'-ATGCTAACCGTAAGAACTTTGCTAAGACTGCTATTACTTTTTATGAAGGATTGGGGTTTT-...
Embodiment 2
[0044] Example 2. Construction of recombinant Pichia pastoris expression vector
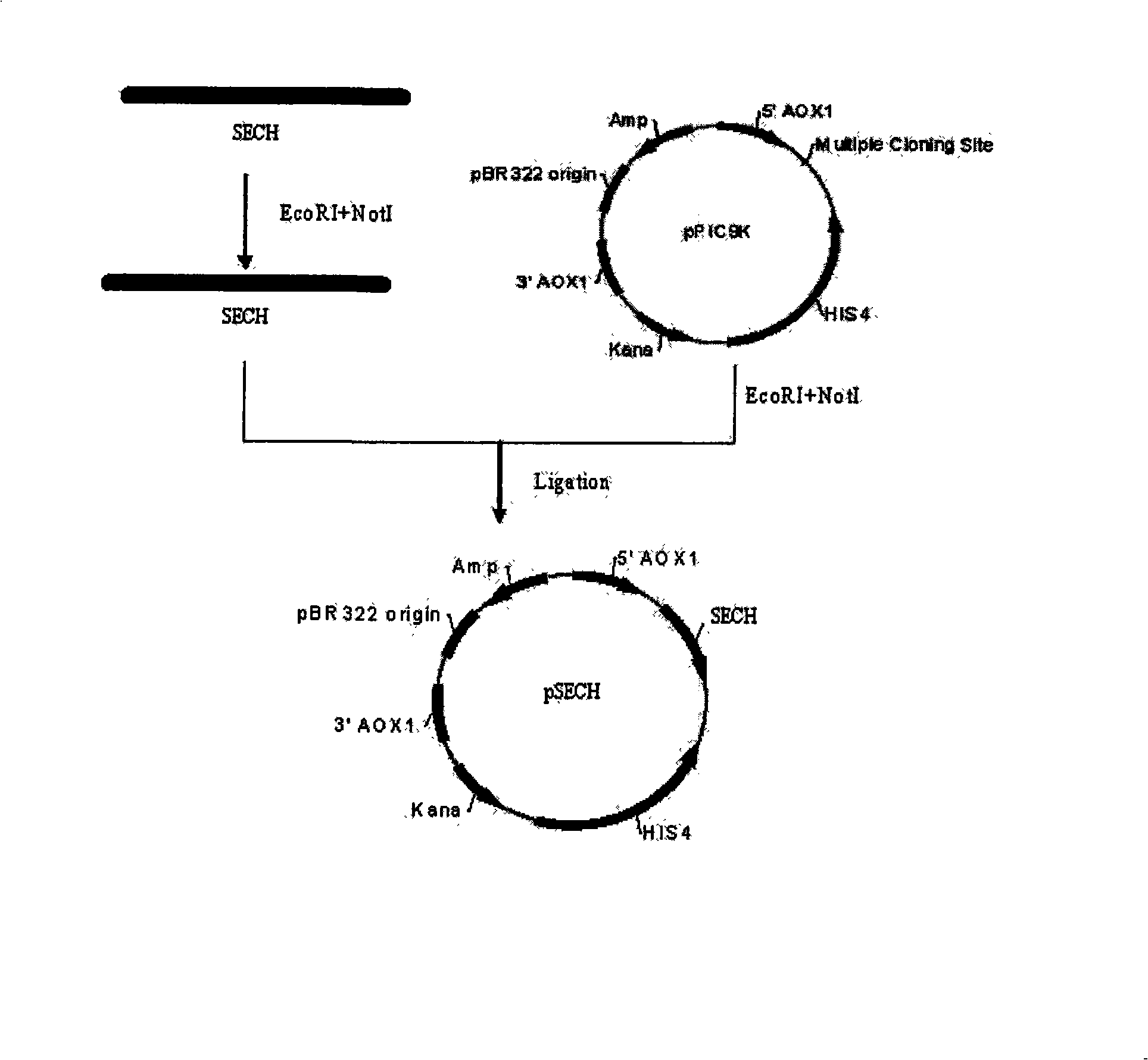
[0045] The PCR amplification product and the original expression plasmid pPIC9K were double-digested with EcoRI and NotI respectively, and the target band was recovered by agarose gel electrophoresis. 4 DNA ligase ligated the recovered target bands to obtain the eukaryotic expression vector pSECH, and used CaCl 2 Transform it into Escherichia coli DH5α by heat shock at 42°C for 90 seconds. Single colonies were screened for Kana resistance. Enzyme digestion identification and DNA sequencing of the selected transformed clones prove that the clones are correct and then sent to Shanghai Bioengineering Co., Ltd. for sequencing. For the specific operation process, see figure 1 .
Embodiment 3
[0046] Example 3. Transformation and screening of recombinant Pichia pastoris
[0047] The identified recombinant expression vector pSECH plasmid DNA was linearized with endonuclease XbaI, and transformed into Pichia pastoris GS115 by electroporation. Immediately after the electric shock, 1ml of pre-cooled 1mol / l sorbitol was added, centrifuged at 3000rpm for 5min, the bacteria were resuspended in 400μl of pre-cooled 1mol / l sorbitol, and 200μl was spread on MD plates (1.34% yeast nitrogen base, 4 ×10 -5 % biotin, 2% glucose, 2% agarose), cultured at 30°C until colonies appeared. Randomly pick bacterial colonies and plant them with different concentrations of antibiotics G418 (0, 0.25mg / ml, 0.50mg / ml, 0.75mg / ml, 1.00mg / ml, 1.50mg / ml, 1.75mg / ml, 2.00mg / ml, 3.00mg / ml, 4.00mg / ml) YPD plate, cultured at 30°C for 2-5 days, and checked the growth of the colony every day. According to the growth situation, the positive clone transformants with high G418 resistance were quickly scre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com