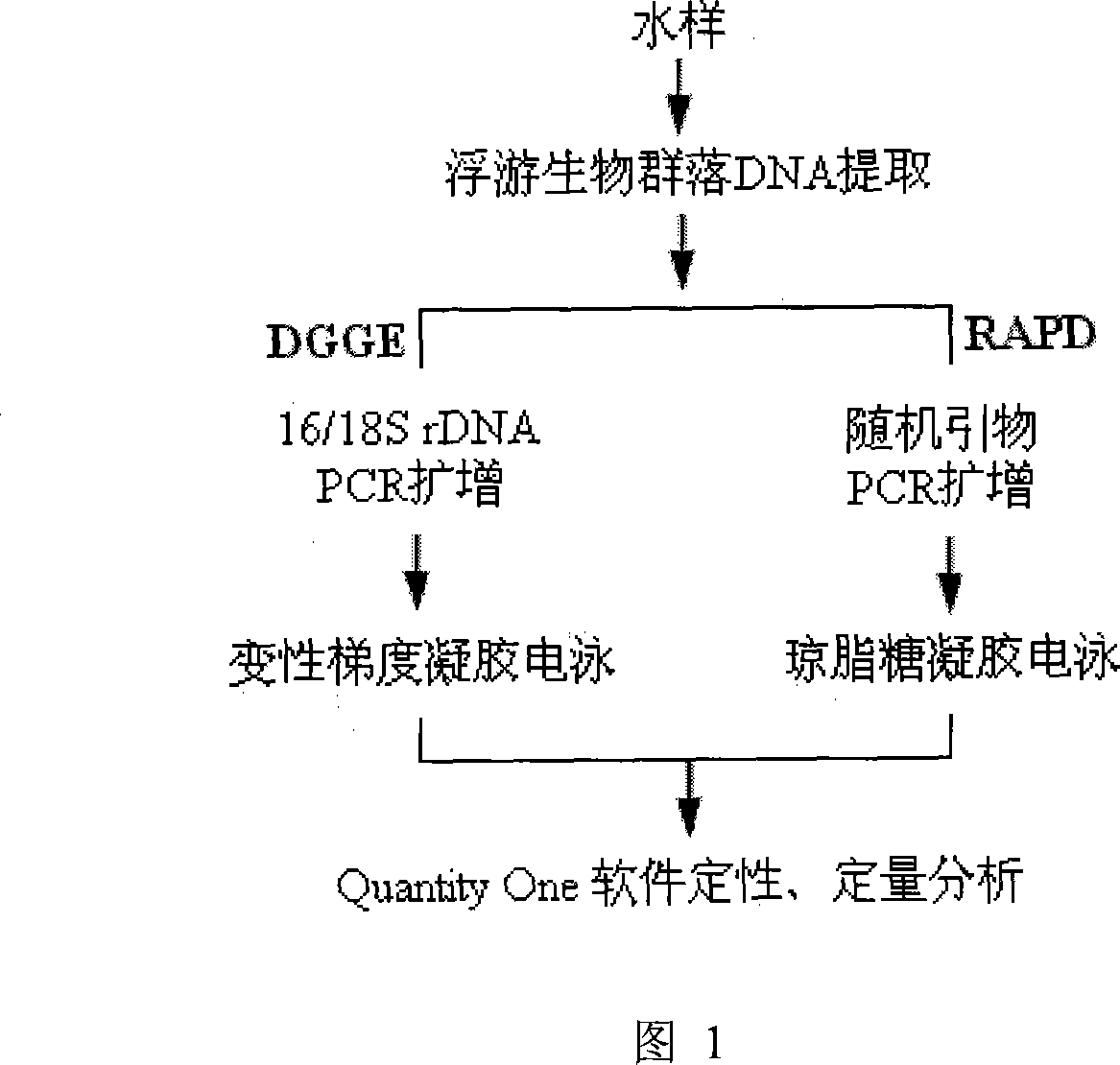
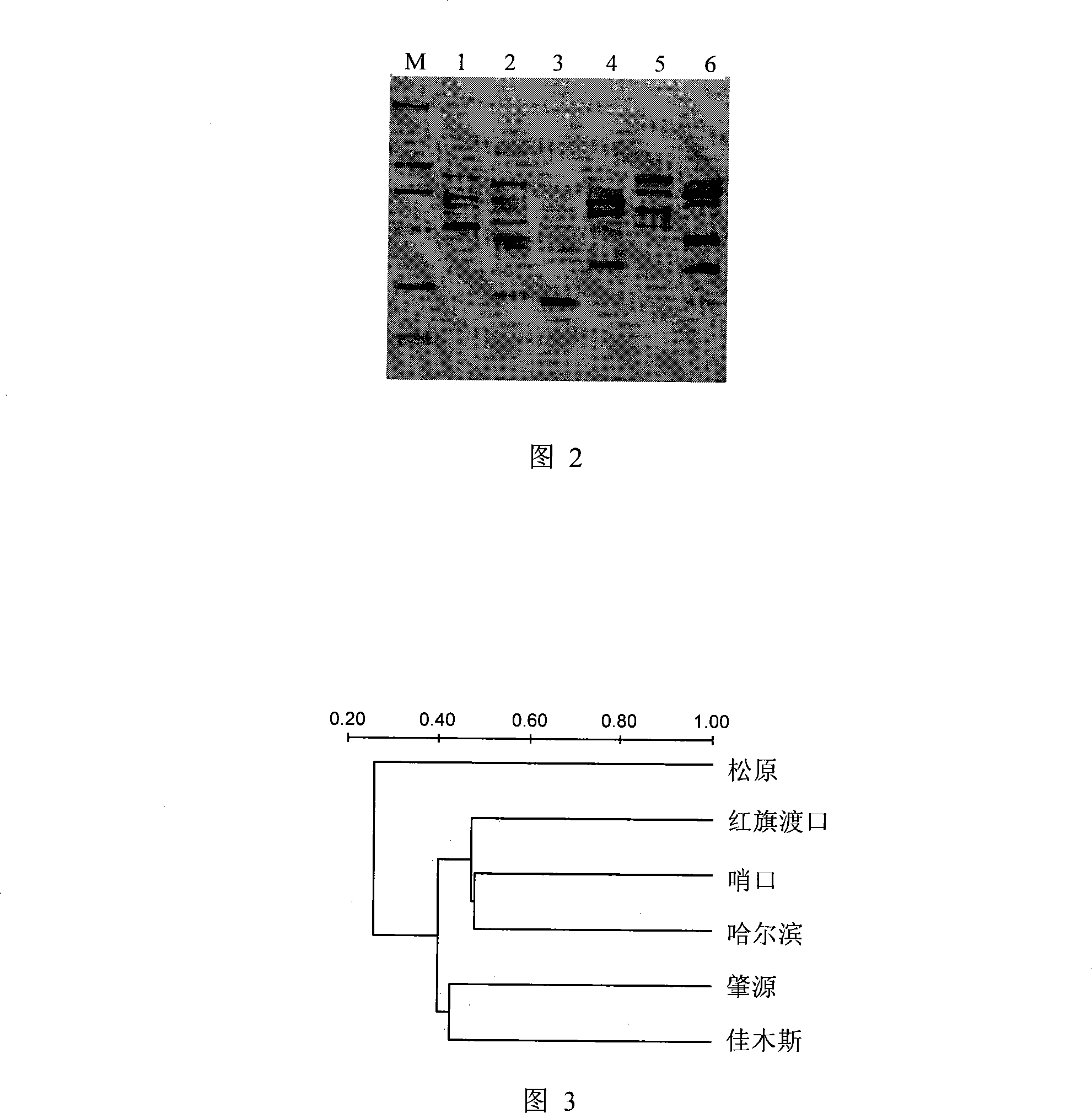
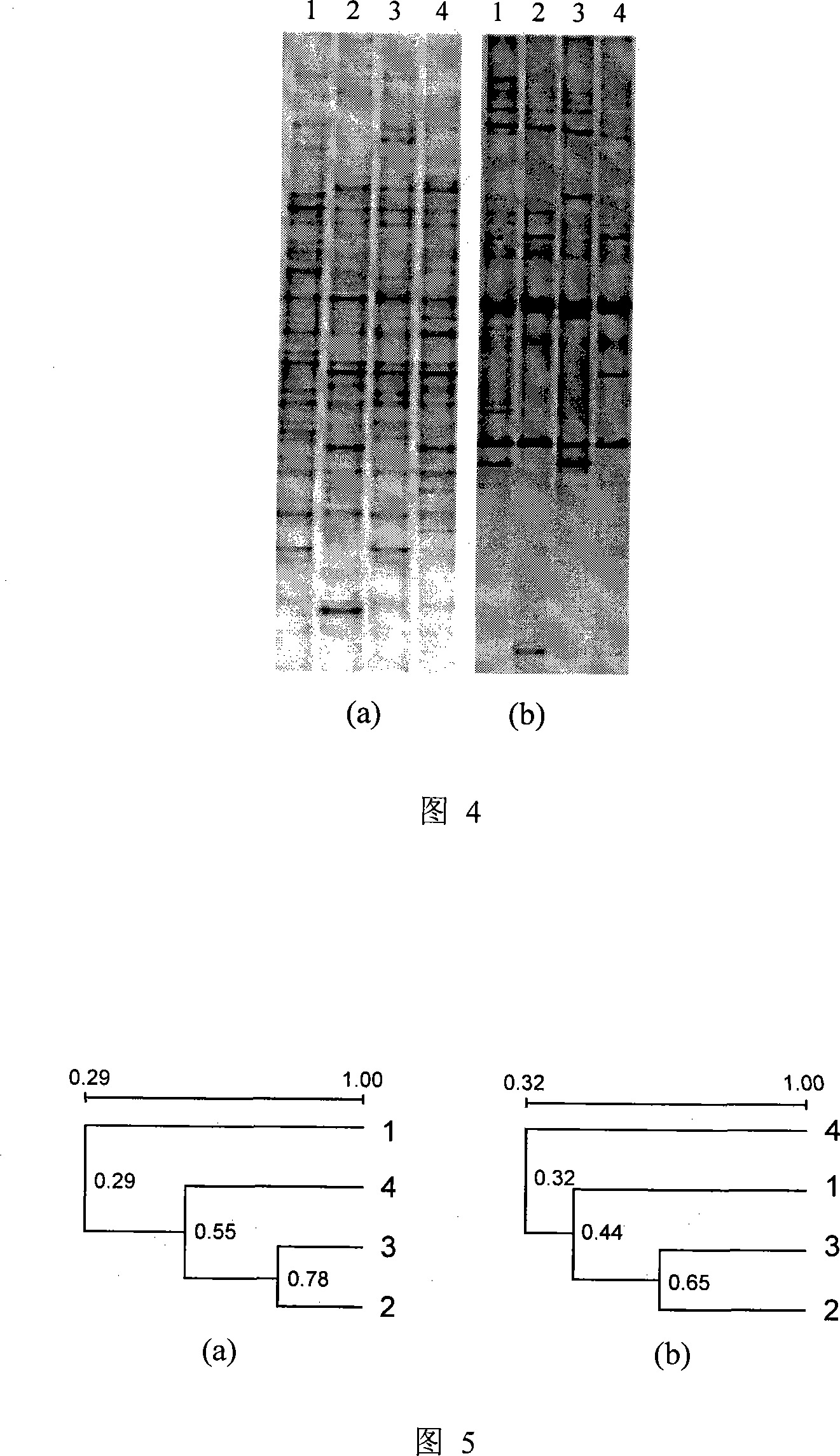
Method for analyzing plankton community DNA polymorphism
A DNA polymorphism, plankton technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of unclear morphological classification characteristics, limited human cognition, small individuals, etc., and achieve the programmed technical route , Comparative analysis of standardized and easy-to-operate effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023]1. Sample Collection A
[0024] Use plexiglass water sampling devices to collect equal volumes of surface and bottom water from 6 sampling points of the Songhua River (Hongqidukou, Shaokou, Songyuan, Zhaoyuan, Harbin, and Jiamusi). After the water samples are fully mixed, use GF / C membrane filter 500mL water sample.
[0025] 2. DNA Extraction of Plankton Community B
[0026] Cut the filtered GF / C filter membrane under sterile conditions (about 1×1mm fragments), and immerse in 3mL lysis buffer (0.5% SDS, 10mM Tris-Cl, 100mM EDTA, 0.1mg / mL proteinase K) in a centrifuge tube, lyse in a water bath at 55°C for 10 h. After centrifugation at 10000r / min for 5min (20°C), transfer the supernatant to a new centrifuge tube, and centrifuge again under the same conditions to obtain the supernatant, which is extracted three times with an equal volume of phenol→phenol:chloroform (1:1)→chloroform. Pre-cooled absolute ethanol (containing 0.3M NaCl) was precipitated at -20°C for 3h. ...
Embodiment 2
[0036] 1. Sample Collection A
[0037] Use plexiglass water collectors to collect equal volumes of surface and bottom water from four indoor ecological simulation boxes (1×1×1m) and mix them. After the water samples are fully mixed, filter 250 mL of water samples with GF / C membranes.
[0038] 2. DNA Extraction of Plankton Community B
[0039] Cut the filtered GF / C filter membrane under sterile conditions (about 1×1mm fragments), and immerse in 3mL lysis buffer (0.5% SDS, 10mM Tris-Cl, 100mM EDTA, 0.1mg / mL proteinase K) in a centrifuge tube, lyse in a water bath at 55°C for 10 h. After centrifugation at 10000r / min for 5min (20°C), transfer the supernatant to a new centrifuge tube, and centrifuge again under the same conditions to obtain the supernatant, which is extracted three times with an equal volume of phenol→phenol:chloroform (1:1)→chloroform. Pre-cooled absolute ethanol (containing 0.3M NaCl) was precipitated at -20°C for 3h. After centrifugation at 13000r / min for 10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com