Use of magnetic bead supported matrix and MS for judging mass spectrometry polypeptide spectrum and protein fingerprint
A protein and magnetic bead technology, applied in the field of protein fingerprinting and protein fingerprinting of biological sample analysis, can solve problems such as unseen, limited gel capacity and sensitivity, and inability to observe molecular weight.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 Significance of Differential Expression of Protein Fingerprints in Biological Samples
[0066] (1) Experimental method
[0067] 1. Materials
[0068] 1. Specimen sources: Serum from 10 normal people and 8 patients with pneumonia.
[0069] 2. Reagents: Protein A and G, human standard serum, human growth hormone, insulin (molecular weight 5807Da), anti-insulin antibody, anti-human serum amyloid A antibody, anti-fibrinopeptide A antibody; acetonitrile, trifluoroacetic acid, SINAPINIC Acid (Sinapinic acid), CHAPS, Urea, NaAC, Tris-HCl were purchased from Sigma or donated.
[0070] 3. Quality control: add human growth hormone (Sigma) to human standard serum (Sigma), and adjust the concentration of human growth hormone in standard human serum to be 50 ng / ml.
[0071] 2. Method
[0072] 1. Collection of samples: After the whole blood is collected, draw the serum and store it at -80°C; take out the serum sample in the refrigerator at -80°C, and put it on the ice b...
Embodiment 2
[0081] Example 2 Sequence identification of serum biomarkers
[0082] Because the biomarkers in this invention are identified by mass spectrometry and magnetic bead support matrix and antibody column, they can be detected by mass spectrometry to directly know their specific identity. This method is more accurate than antibody-based ELISA and immunofluorescence methods.
[0083] The serum 1536.7Da biomarker protein was reduced from a fibrinopeptide A antibody column using MALDI-Qq-TOF mass spectrometry (MS / MS), post-source fragmentation (PSD) and protein ladder sequencing Sort. By breaking molecules into pieces, protein ladders can be generated. This gradient is then analyzed by mass spectrometry. The biomarker and its chemical structure were identified as fibrinopeptide A (arranged from N-terminus to C-terminus):
[0084] N-terminal Ala-Asp-Ser-Gly-Glu-Gly-Asp-Phe-Leu-Ala-Glu-Gly-Gly-Gly-Gly-Val-Arg C-terminal
[0085] Check the chemical structure of the normal fibrinopep...
Embodiment 3
[0088] Example 3 Normal Human Serum Protein Fingerprint and Mass Spectrometry Peptide Spectrum
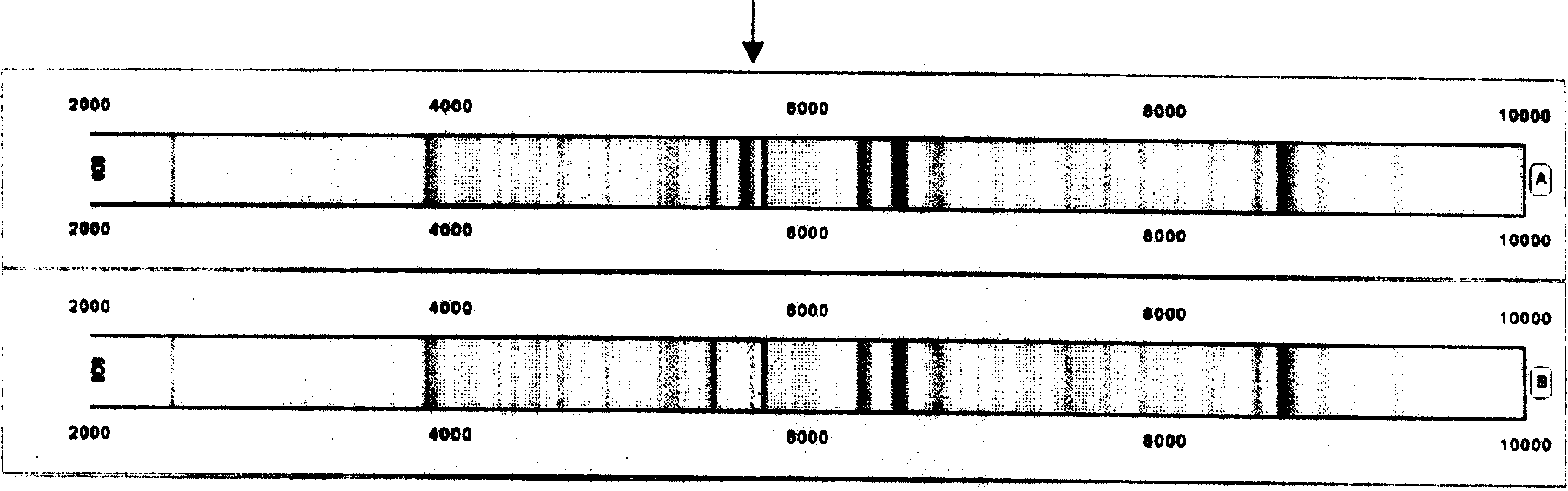
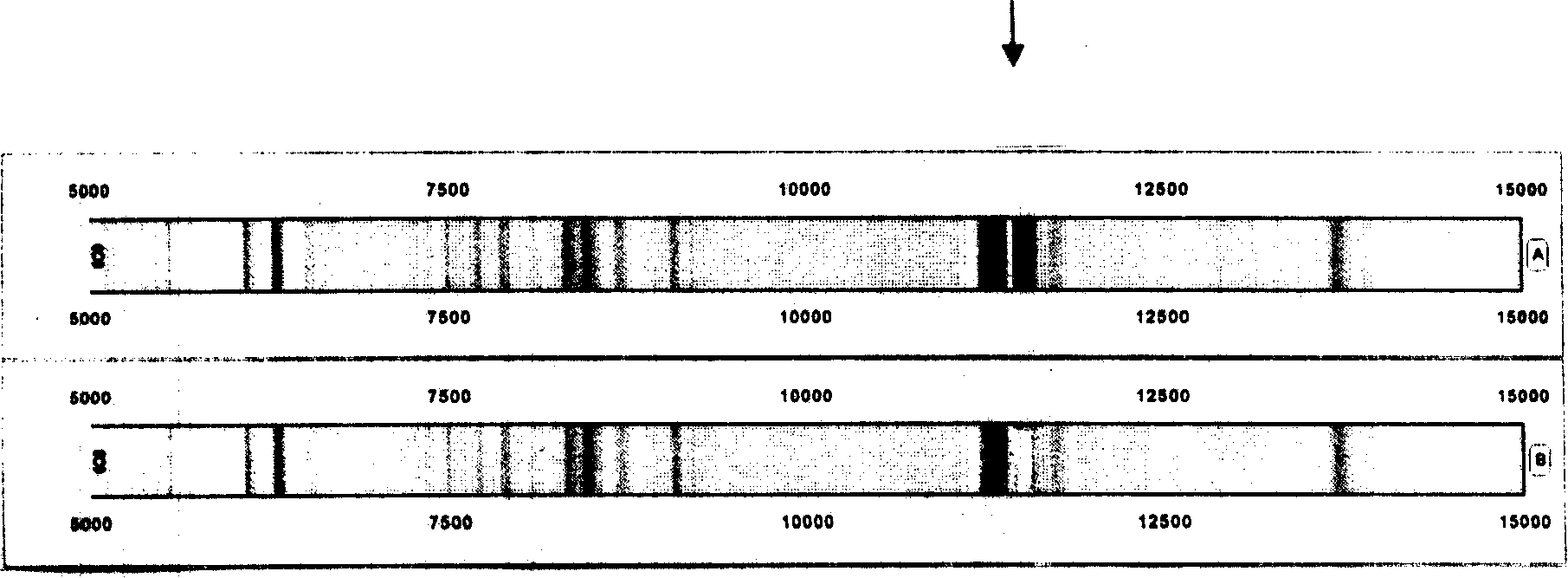
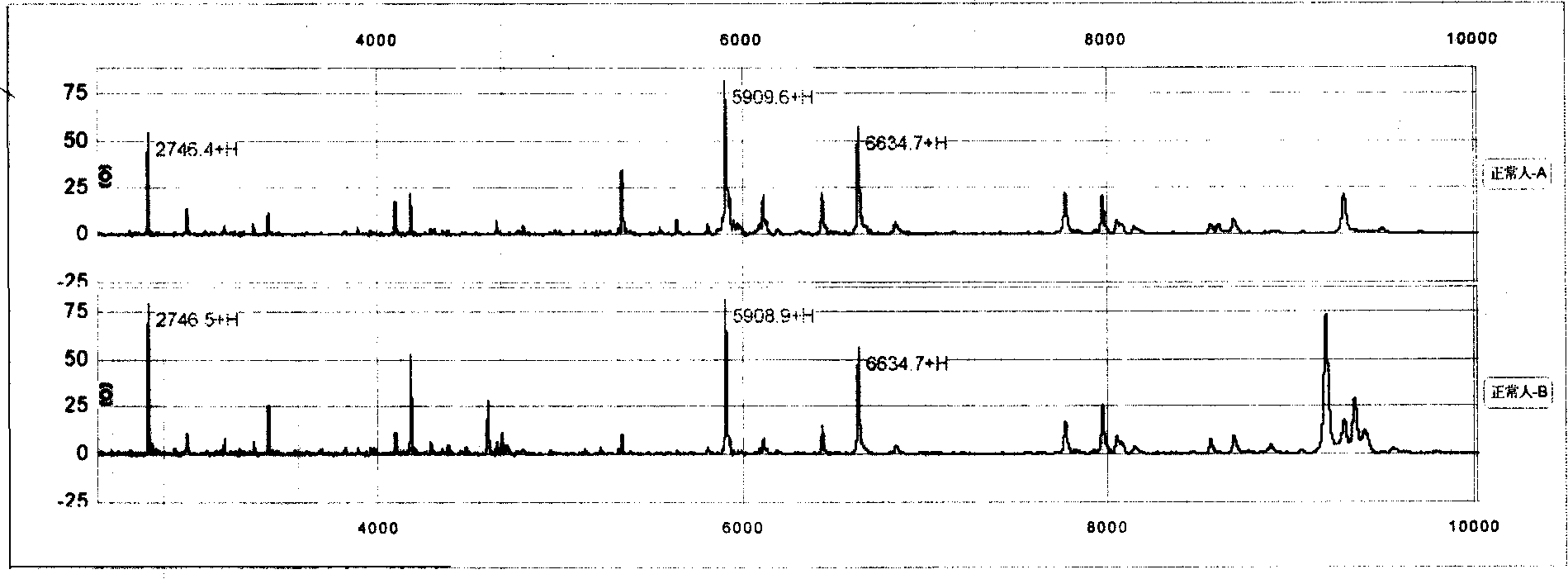
[0089] Quantitative mass spectrometry regulation: Before each test, use the standardized quality control serum of mass spectrometry to adjust the standard peak 6634.0Da equal intensity for quantification in the standardized quality control serum to the maximum value of 40-50% signal intensity ( image 3 ). image 3 The fingerprints and mass spectrometry peptide spectra of two serum proteins of normal people are shown. 2746±1Da, 5909±1Da, 6634±1Da peaks can be used as standard peaks for molecular weight quality control.
[0090] in conclusion
[0091] Protein fingerprinting of biological samples captured by magnetic bead-supported matrix for mass spectrometry analysis - in a computer-readable barcode format (protein fingerprint). The present invention provides for determining the presence of a difference between a first and a second sample of a biological sample. Including A) dete...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com