Biomarkers and uses thereof in prognosis and treatment strategies for right-side colon cancer disease and left-side colon cancer disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of RT-PCR Primer-Probes that Measure in FFPE Tissue the mRNA Species Targeted by the ap-Colon Microarray Probes
[0108]mRNA will be extracted from a number of colon cancer cell lines as well as from paraffin (FFPE) blocks prepared from these cell lines. This will enable direct assessment of the probes in the FFPE material and comparison with the “fresh state”. Initial assessment will be performed using 13 different assay primer-probes pairs (8 from ap-Colon (two per gene) and 5 normalization controls). All assays will be performed in triplicate. The probes will be verified as providing comparable results in fresh tissues (cell lines) and matched FFPE counterparts. Quantitative RT-PCR with ΔΔCT methods for data analysis will be used to assess the utility of the probes. If suitable primer-probes cannot be found for the initial choice of genes, the list will be screened to identify replacement genes found in the development of ap-Colon. The RL-COLON pair of tests will use ...
example 2
Materials and Methods
[0113]The present example is provided to present the various materials, methods and statistical tools employed in the development and practice of the present invention.
[0114]Statistical analysis. The language R http: / / www.r-project.org / was used for all statistical analyses. Survival models were fit with the R package survival. The microarray annotation package hgu133plus2.db in BioConductor http: / / www.bioconductor.org / was also used. The proportional hazard condition was verified with the cox.zph function. All p-values in survival models refer to the p-value of the logrank score of a Cox proportional hazard model (CPH). A CPH is considered statistically significant if the p-value of the logrank score is <0.05.
[0115]Microarray dataset of colon cancer samples. In the present examples, the Gene Expression Omnibus (http: / / www.ncbi.nim.nih.gov / gds) data series GSE14333 was used.13 The characteristics of the data series GSE14333 are provided in Table 323.
[0116]Sample...
example 3
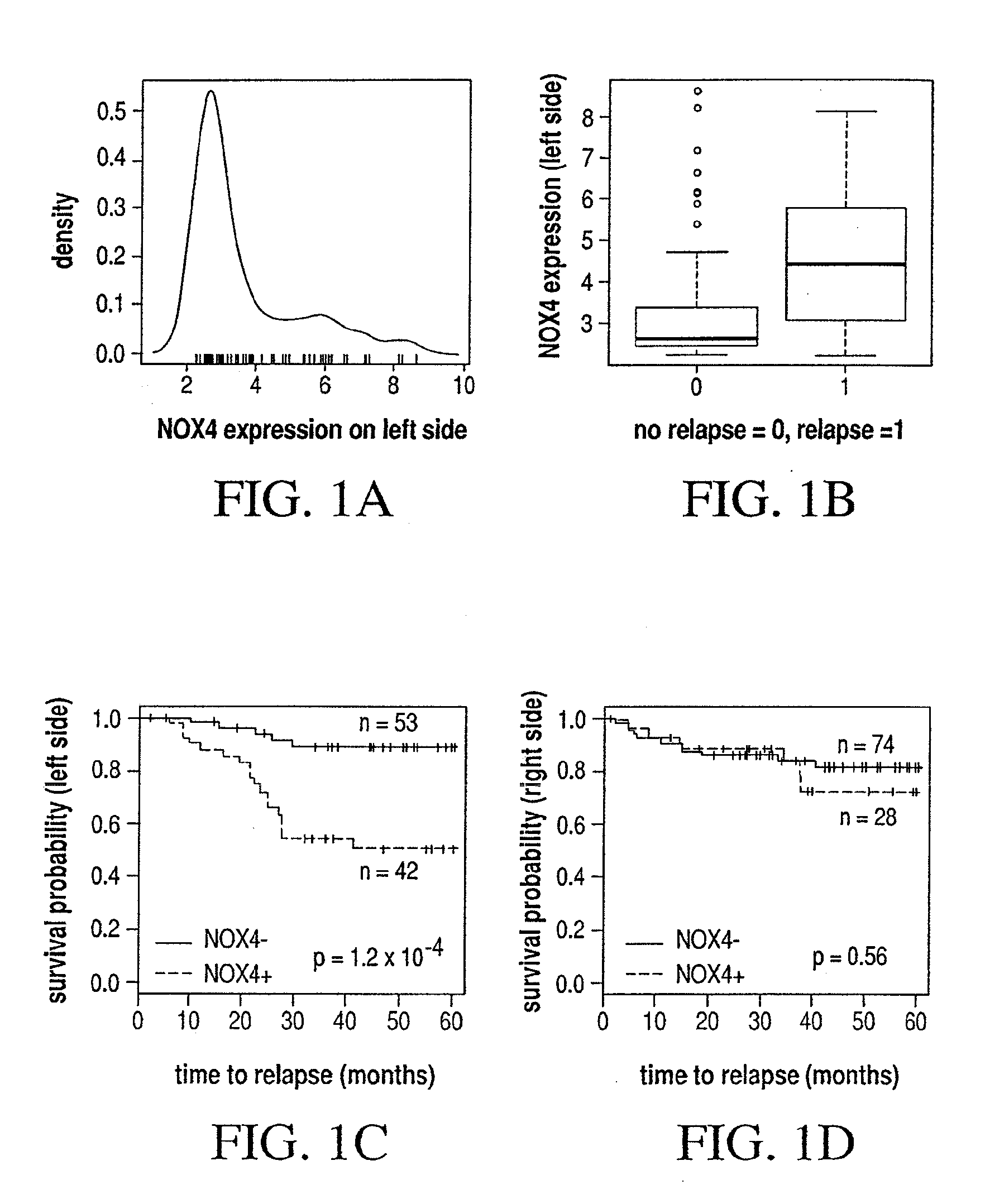
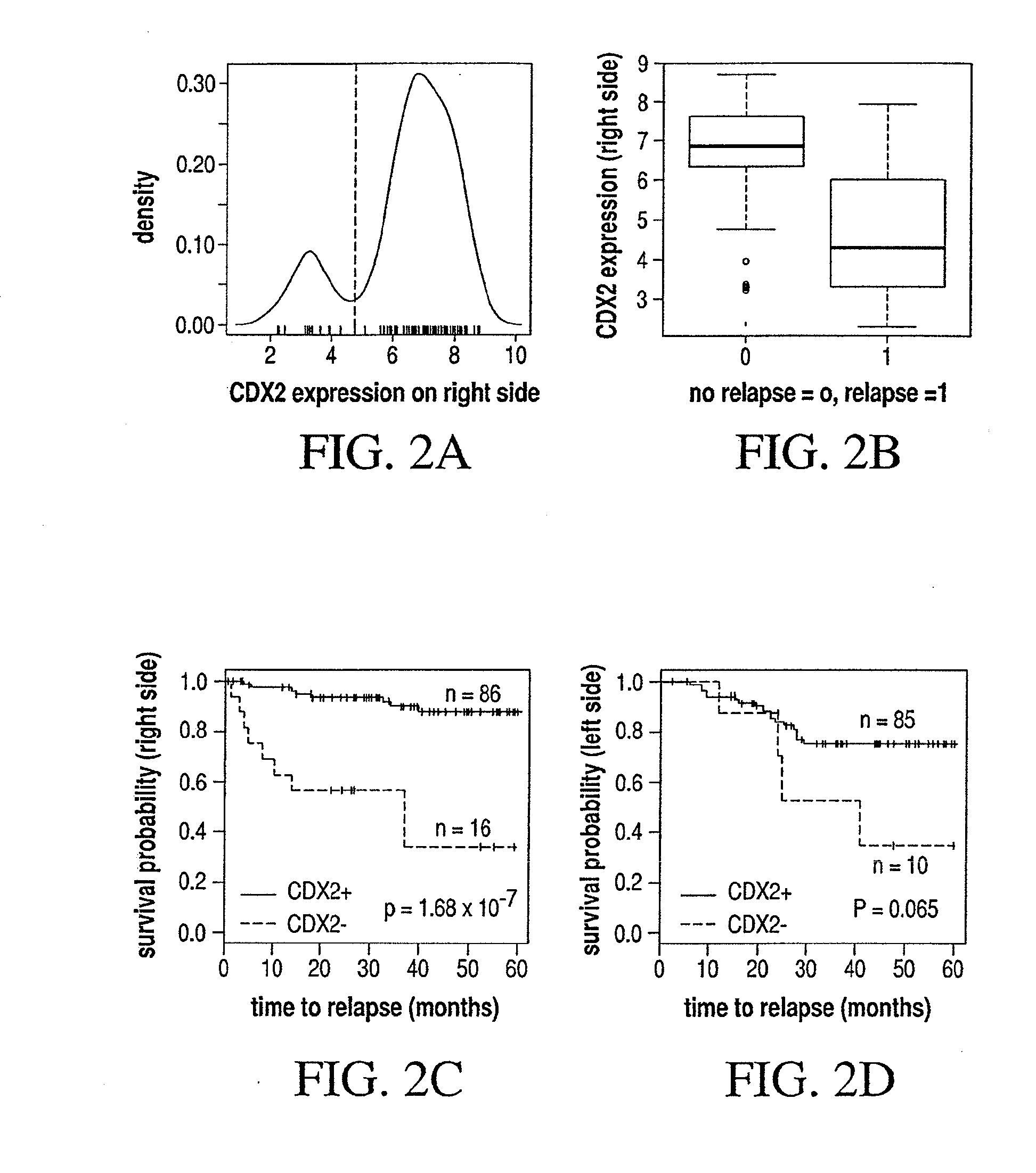
Different Pathways dominate Progression to Relapse in LCC and RCC
[0128]The present example demonstrates the location specificity of the dominant pathway to relapse in colon cancer. Attention is focused on samples in GSE14333 with Dukes stage A, B or C. Table 3 demonstrates the characteristics of patients in GSE14333.
TABLE 3Characteristics of patients in GSE14333relapsechemoin stagein stageDukes stagegenderA, B, CA, B, Cno.(A / B / C / D)(M / F)(no / yes)(no / yes)all tumors29044 / 94 / 91 / 61164 / 126180 / 46 142 / 87 left side12218 / 37 / 40 / 2777 / 4570 / 2355 / 40right side12517 / 44 / 41 / 2359 / 6684 / 1763 / 39rectum398 / 12 / 10 / 926 / 1324 / 6 22 / 8 other41 / 1 / 0 / 22 / 22 / 02 / 0
TABLE 4Genes and associated pathways most significantly implicated in relapse in leftside colon cancer with Dukes stage A, B or C. Left side:CPHdirectionpathwaysmultistateprobegenep-valuein relapseeffected*marker236028_atIBSP2.7 × 10−5UPFANOX4210095_s_atIGFBP31.0 × 10−4UPP53NOX4213425_atWNT5A2.5 × 10−4DOWNWNTMMP3223121_s_atSFRP23.1 × 10−4UPWNTNOX4229271_x_atCOL11...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Angle | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com