Use of proteases for gluten intolerance
a technology of proteases and gluten, applied in the direction of peptide/protein ingredients, drug compositions, metabolic disorders, etc., can solve the problems of inability of the intestine to absorb nutrients, negative test results, and inconclusive results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
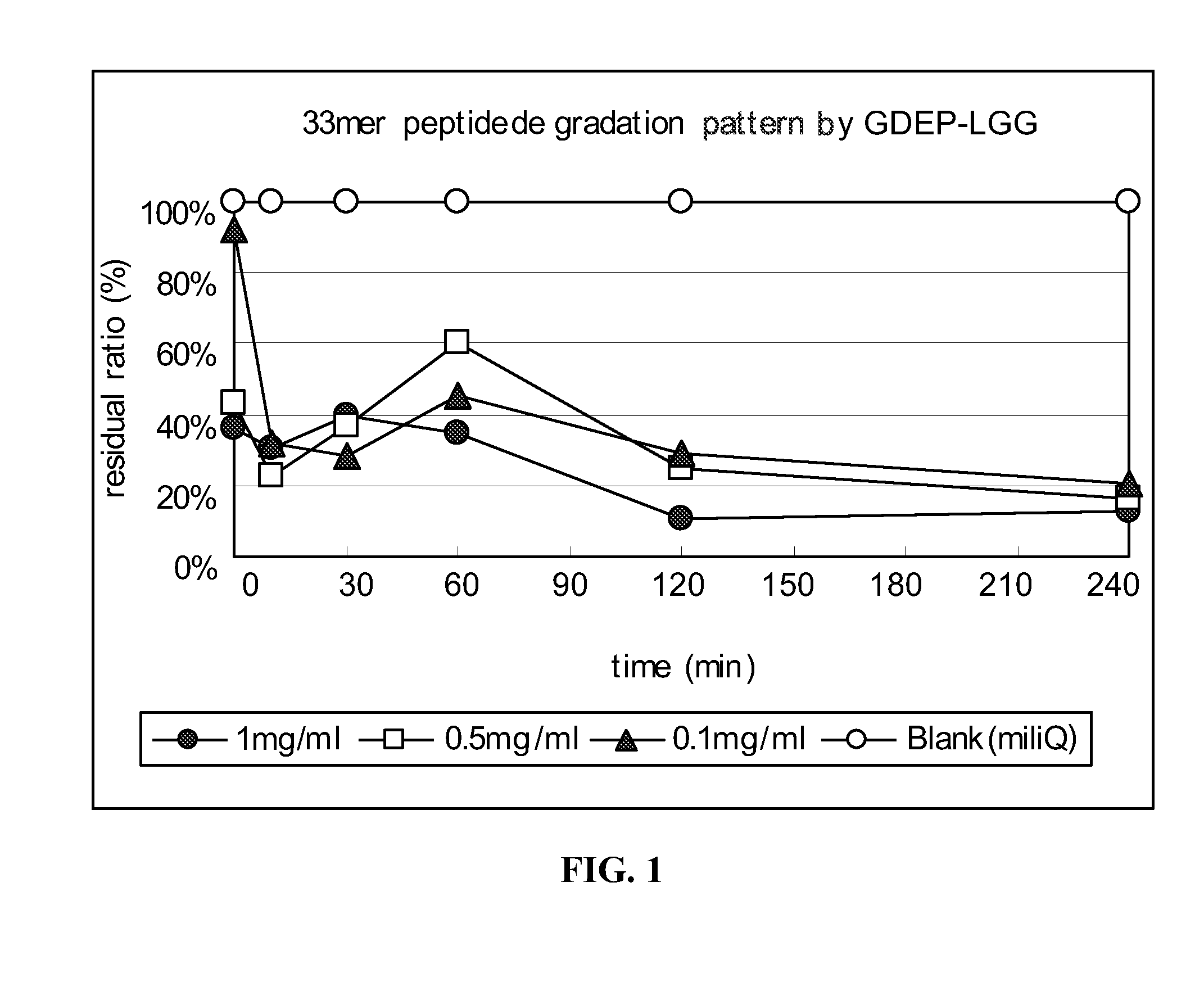
33-mer Peptide Degradation by GDEP-LGG
[0137]A 33-mer peptide (1 mg / ml; LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF (SEQ ID NO:1)) was incubated with GDEP-LGG at various concentrations (0 mg / ml; 0.1 mg / ml, 0.5 mg / ml, and 1.0 mg / ml) in the presence of simulated intestinal fluid (SIF; pH 6.8) from U.S. Pharmacopeia. A 30 μL reaction volume comprised 10 μL 33-mer peptide, 10 μL enzyme solution, and 10 μL 3× SIF buffer. The reactions were carried out for about 15, 30, 60, 120, and 240 minutes at about 37° C. and, then, 5 min at 90° C. to stop the enzyme reaction.
[0138]Samples of enzyme-treated 33-mer were analyzed by enzyme-linked immunosorbent assay (“ELISA”) using a HRP-conjugated polyclonal antibody developed against the 33-mer peptide.
[0139]The results of the time-course and dose-response analysis of 33-mer peptide degradation by GDEP-LGG are shown in FIG. 1. Incubation with GDEP-LGG (at all concentrations) for about 120 minutes reduced the antigenicity of the 33-mer by at least about 70%.
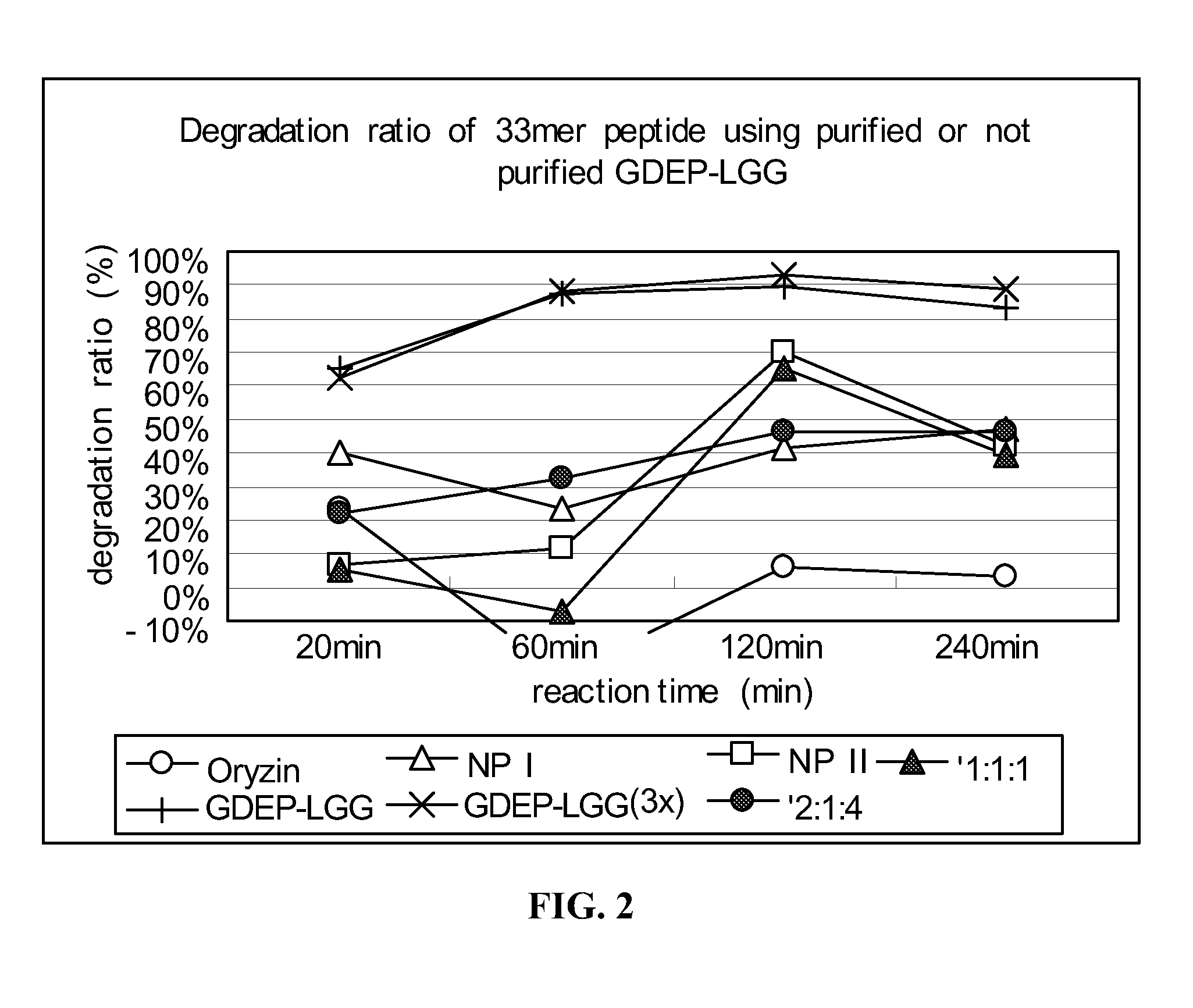
example 2
33-mer Peptide Degradation by Components of GDEP-LGG
[0140]Individual components of GDEP-LGG, including Oryzin, NP I, and NP II, were tested for enzymatic activity against the 33-mer. The 33-mer peptide (1 mg / ml) was incubated with 0.1 mg / ml enzyme solution comprising GDEP-LGG; 3X GDEP-LGG (i.e., 0.3 mg / ml); Oryzin; NP I; NP II; or combinations of Oryzin, NP I, and NP II (at ratios of 1:1:1 and 2:1:4) in the presence of simulated intestinal fluid (SIF; pH≈6.8). A 30 μL reaction volume comprised 10 μl 33-mer peptide, 10 μL enzyme solution, and 10 μL 3× SIF buffer. The reactions were carried out for about 20, 60, 120, and 240 minutes at about 37° C. and, then, 5 min at 90° C. to stop the enzyme reaction.
[0141]Samples of enzyme-treated 33-mer were analyzed as in Example 1. Degradation of the 33-mer by GDEP-LGG and its individual component enzymes is shown in FIG. 2. Incubation with GDEP-LGG (at both concentrations tested) for about 60 minutes resulted in degradation of about 90% of the ...
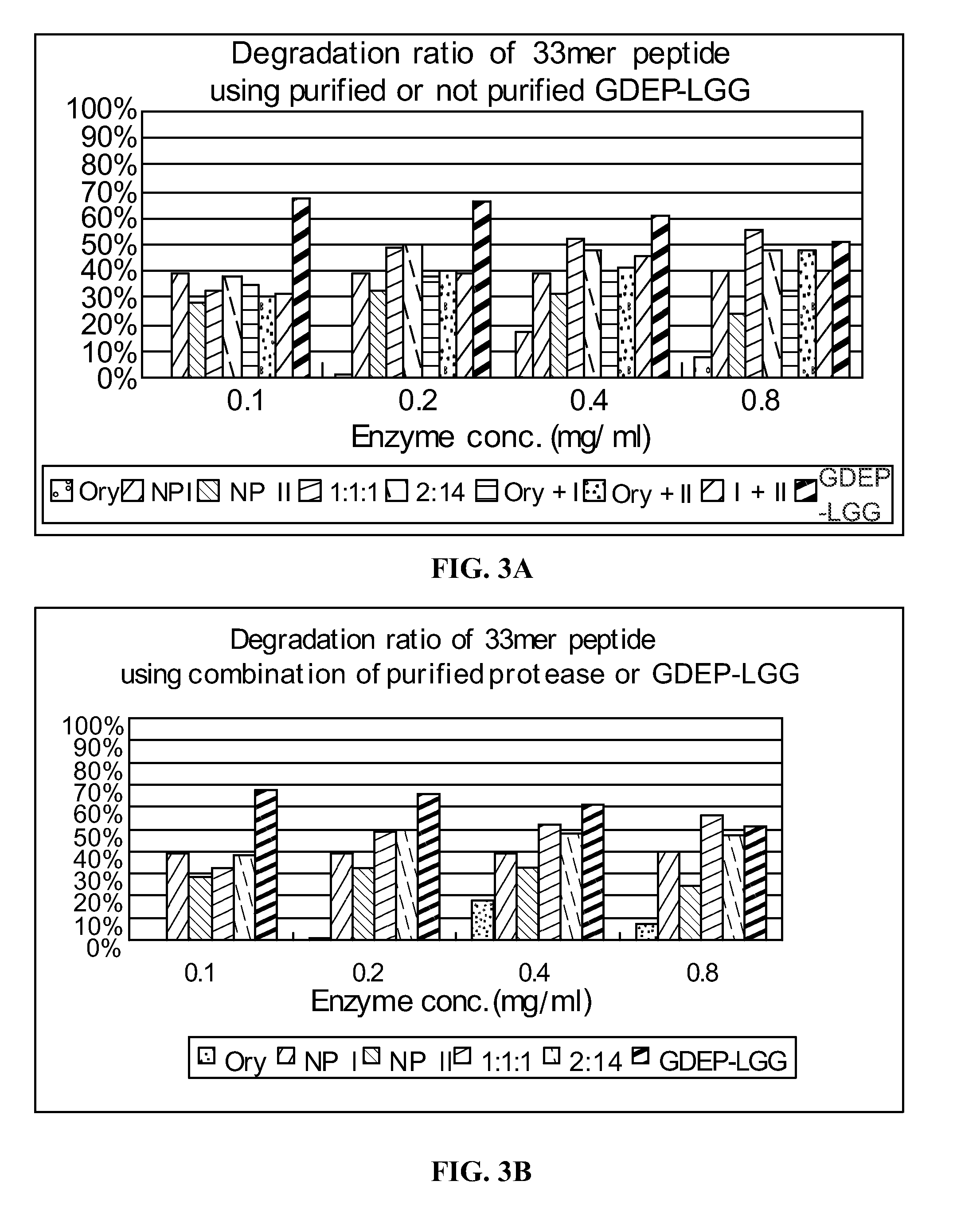
example 3
Comparison of GDEP-LGG and Other Digestive Proteases
[0144]The 33-mer peptide (1 mg / ml) was incubated with 0.25 mg / ml enzyme solution (containing pepsin, proteinase K, or GDEP-LGG) in the presence of simulated intestinal fluid (SIF; pH≈6.8) or simulated gastric fluid (SGF; pH≈2.0). A 40 μL reaction volume comprised 10 μL 33-mer peptide, 10 μL enzyme solution, and 20 μL 2× SIF or SGF buffer. The reactions were carried out for about 15 hours at 37° C. and, then, 5 min at 90° C.
[0145]Samples of enzyme-treated 33-mer were analyzed by reverse phase high pressure liquid chromatography (“HPLC”) using an Agilent 1100 HPLC system. Samples were loaded onto an Eclipse SB-C18 (ID2.1 mm×150 mm, 3.5 um) column. The mobile phase comprised of (A) distilled water with 0.1% formic acid and (B) acetonitril with 0.1% formic acid was used to elute targets in gradient mode (0 min, 2% B→10 min, 25% B→40 min, 40% B→45 min, 100% B→50 min, 100% B). Flow rate was set at 0.2 mL / min and the injection volume was ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com