Methods and means for obtaining modified phenotypes
phenotype technology, applied in the field of reducing the phenotypic expression of a nucleic acid sequence of interest in eucaryotic cells, can solve the problems of difficult detection of the same plant and easy virus transmission
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
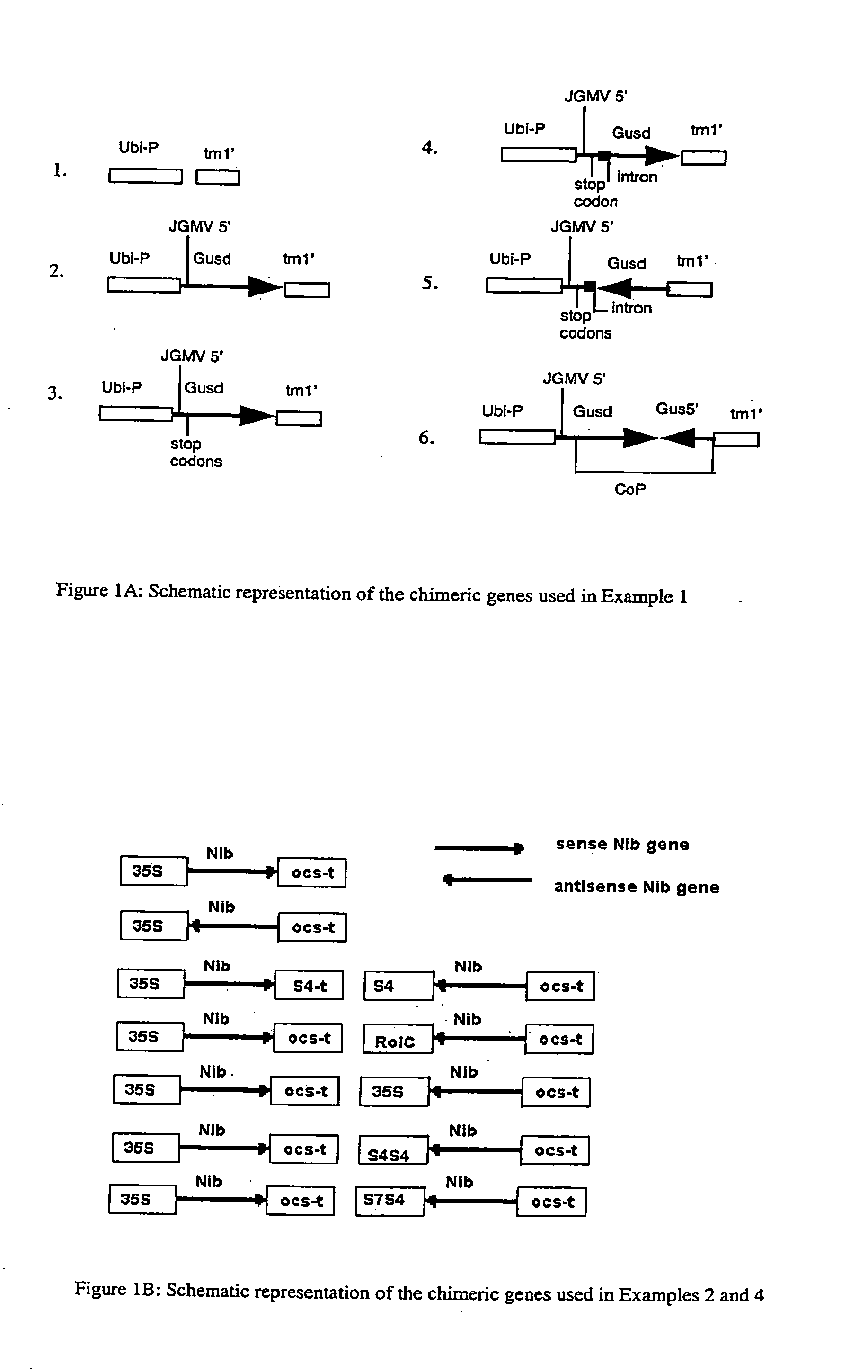
Comparison of Chimeric Genes Comprising Only Antisense, Only Sense, or Both Sense and Antisense (Complimentary Pair (CoP)) Sequence for Reduction in Phenotypic Expression of an Integrated β-Glucuronidase Gene
[0174] Transgenic rice tissue expressing β-glucuronidase (GUS) from a single transgene (and hygromycin resistance from a hph gene) (lines V10-28 and V10-67) was supertransformed using vectors that contained the bar gene conferring phosphinothricin resistance and various sense, antisense and CoP constructs (see FIG. 1A) derived from a crippled GUS (GUSd) gene. The supertransformed tissue was maintained on hygromycin and bialaphos selection media for 3 weeks then analyzed for GUS activity. A crippled GUS gene was used so that expression from this gene would not be superimposed on the endogenous GUS activity.
[0175] The figures in Table 2 represent the rate of MU production measured by absorption at 455 nm, with excitation at 365 nm of 1.5 μg of total protein in a reaction volume ...
example 2
Comparison of the Efficiency of Using Chimeric Genes Comprising Only Antisense Genes, Only Sense Genes, or Both Genes Simultaneously for Obtaining Virus Resistance in Transgenic Plants
[0178] Gene constructs were made using the PVY protease encoding sequence SEQ ID No 1) in a sense orientation, an antisense orientation and in a complimentary pair (CoP) orientation, where the T-DNA comprised both the sense and antisense chimeric genes each under control of their own promoter. In all three arrangements the CaMV35S promoter was used. Five different versions of CoP constructs were made in which the second promoter was either the CaMV35S promoter, the S4 promoter, the double S4 promoter, the S7 enhanced S4 promoter, or the vascular specific rolC promoter (see FIG. 1B).
[0179] These constructs were transformed into tobacco (via Agrobacterium mediated DNA transfer) and approximately 25 independently transformed plants were recovered per chimeric gene construct. The transgenic plants were t...
example 3
Inheritance of Extreme Resistance in Plants from Example 2
[0182] Plants from Example 2 were allowed to self-fertilize and their seeds were collected. Seeds originating from plants showing extreme resistance and low transgene copy number for CoP constructs 35S-Nia / S4-AntisenseNia and 35S-Nia / 35S-AntisenseNia, and seeds from the sense and the antisense plants showing extreme resistance, were germinated and grown in the glasshouse. Plants were also grown from seed collected from two susceptible CoP lines, two susceptible sense gene only lines and two susceptible antisense gene only lines. Twenty plants from each line were selected for overall uniformity of size and development stage, put into individual pots, allowed to recover for one week, then inoculated with PVY. The plants were scored for virus symptoms 2, 4, and 7 weeks after inoculation. The results (Table 5) showed that all eight plant lines of 35S-Nia / S4-antisenseNia and 35S-Nia / 35S-antisenseNia containing one or two gene cop...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| fatty acid profile | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com