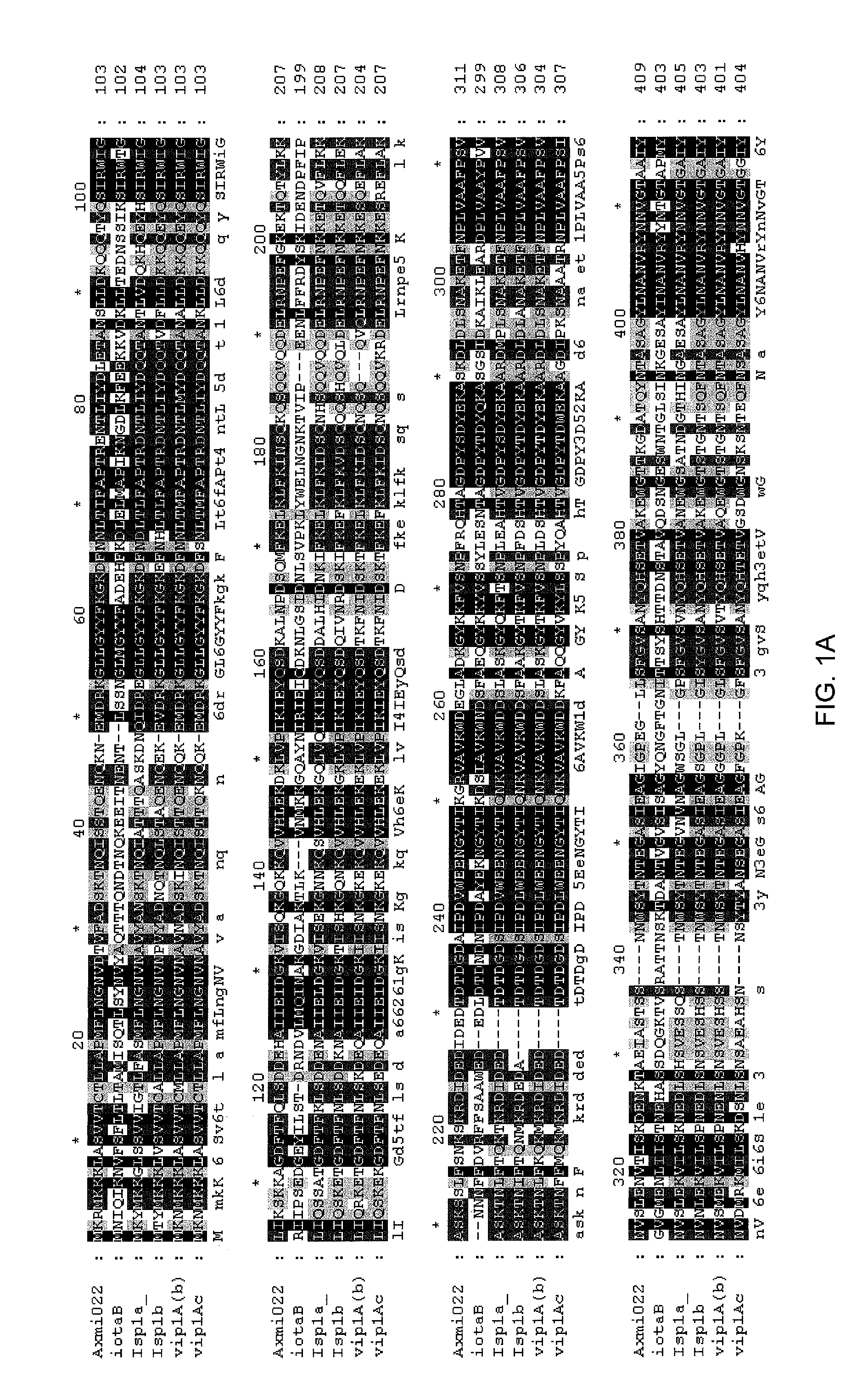
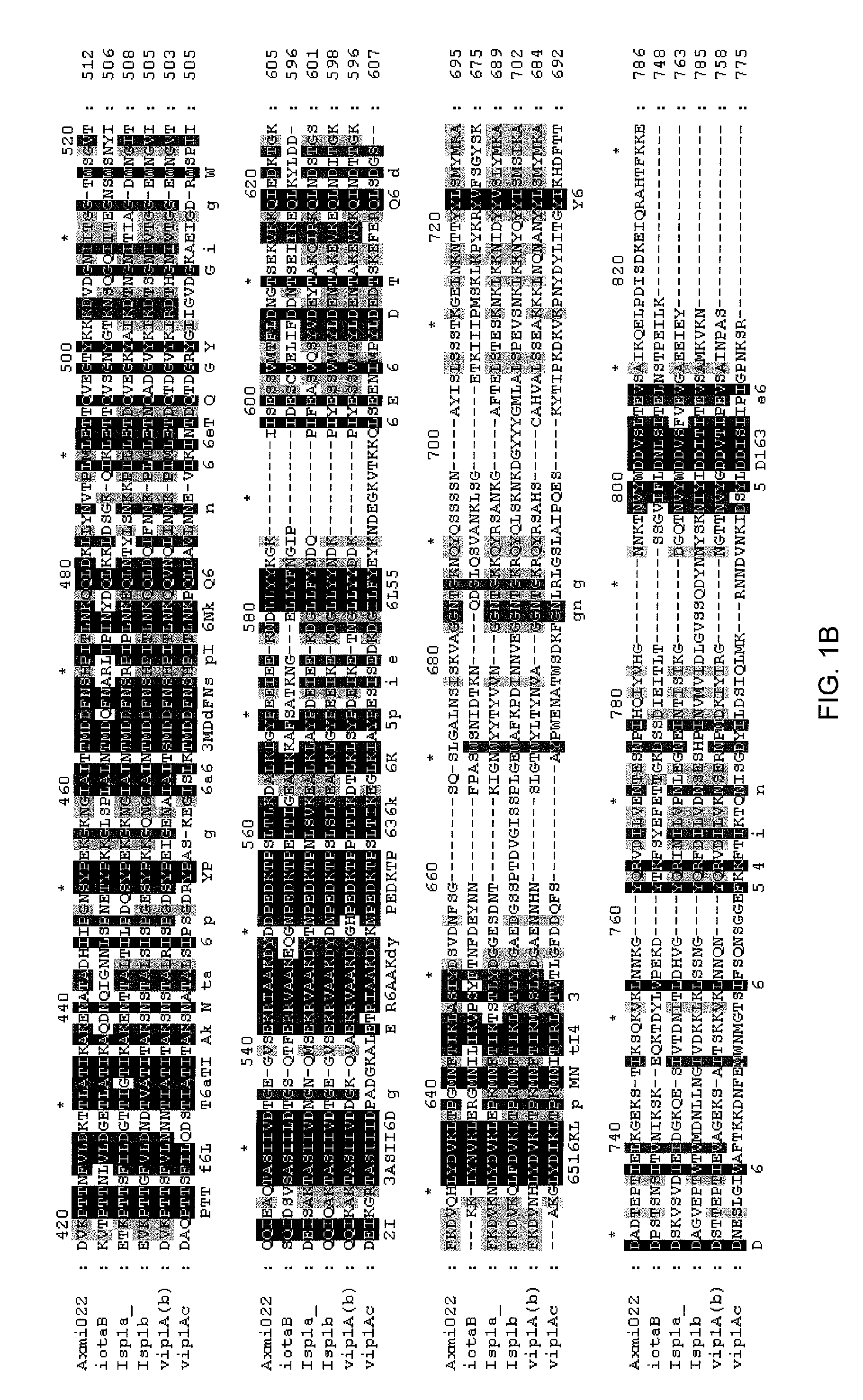
Family of pesticidal proteins and methods for their use
a technology of pesticidal proteins and family, applied in the field of molecular biology, can solve problems such as larval death, and achieve the effect of improving yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0105] Strains ATX14759, ATX14875, ATX13026, ATX13002, ATX9387, ATX13045, ATX21738, ATX14833, ATX1489, ATX15398 and ATX12972 were selected for analysis. Pure cultures of each strain were grown in large quantities of rich media. The cultures were centrifuged to harvest the cell pellet. The cell pellet was then prepared by treatment with SDS by methods known in the art, resulting in breakage of the cell wall and release of DNA. Proteins and large genomic DNA were then precipitated by a high salt concentration. The plasmid DNA was then precipitated with ethanol. In several instances, the plasmid DNA was separated from any remaining chromosomal DNA by high-speed centrifugation through a cesium chloride gradient. Alternatively, the plasmid DNA was purified by binding to a resin, as known in the art. For each strain, the quality of the DNA was checked by visualization on an agarose gel by methods known in the art.
example 2
Cloning of Genes
[0106] DNA libraries were prepared from the plasmid DNA or each strain. This may be achieved in many ways as known in the art. For, example, the purified plasmid DNA can be sheared into 5-10 kb sized fragments and the 5′ and 3′ single stranded overhangs repaired using T4 DNA polymerase and Klenow fragment in the presence of all four dNTPs, as known in the art. Phosphates can then be attached to the 5′ ends by treatment with T4 polynucleotide kinase, as known in the art. The repaired DNA fragments can then be ligated overnight into a standard high copy vector (i.e. pBLUESCRIPT™ SK+), suitably prepared to accept the inserts as known in the art (for example by digestion with a restriction enzyme producing blunt ends).
[0107] The quality of the resulting DNA libraries was analyzed by digesting a subset of clones with a restriction enzyme known to have a cleavage site flanking the cloning site. A high percentage of clones were determined to contain inserts, ideally with ...
example 3
High Throughput Sequencing of Library Plates
[0108] Once the DNA library quality was checked and confirmed, colonies were grown in a rich broth in 2 ml 96-well blocks overnight at 37° C., typically at a shaking speed of 350 rpm. The blocks were centrifuged to harvest the cells to the bottom of the block. The blocks were then prepared by standard alkaline lysis prep in a high throughput format.
[0109] The end sequences of clones from this library were then determined for a large number of clones from each block in the following manner: The DNA sequence of each clone chosen for analysis was determined using the fluorescent dye terminator sequencing technique (Applied Biosystems), by methods known in the art using an automated DNA sequencing machine, and standard oligonucleotide primers that anneal to the plasmid vector in the region flanking the insert.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com