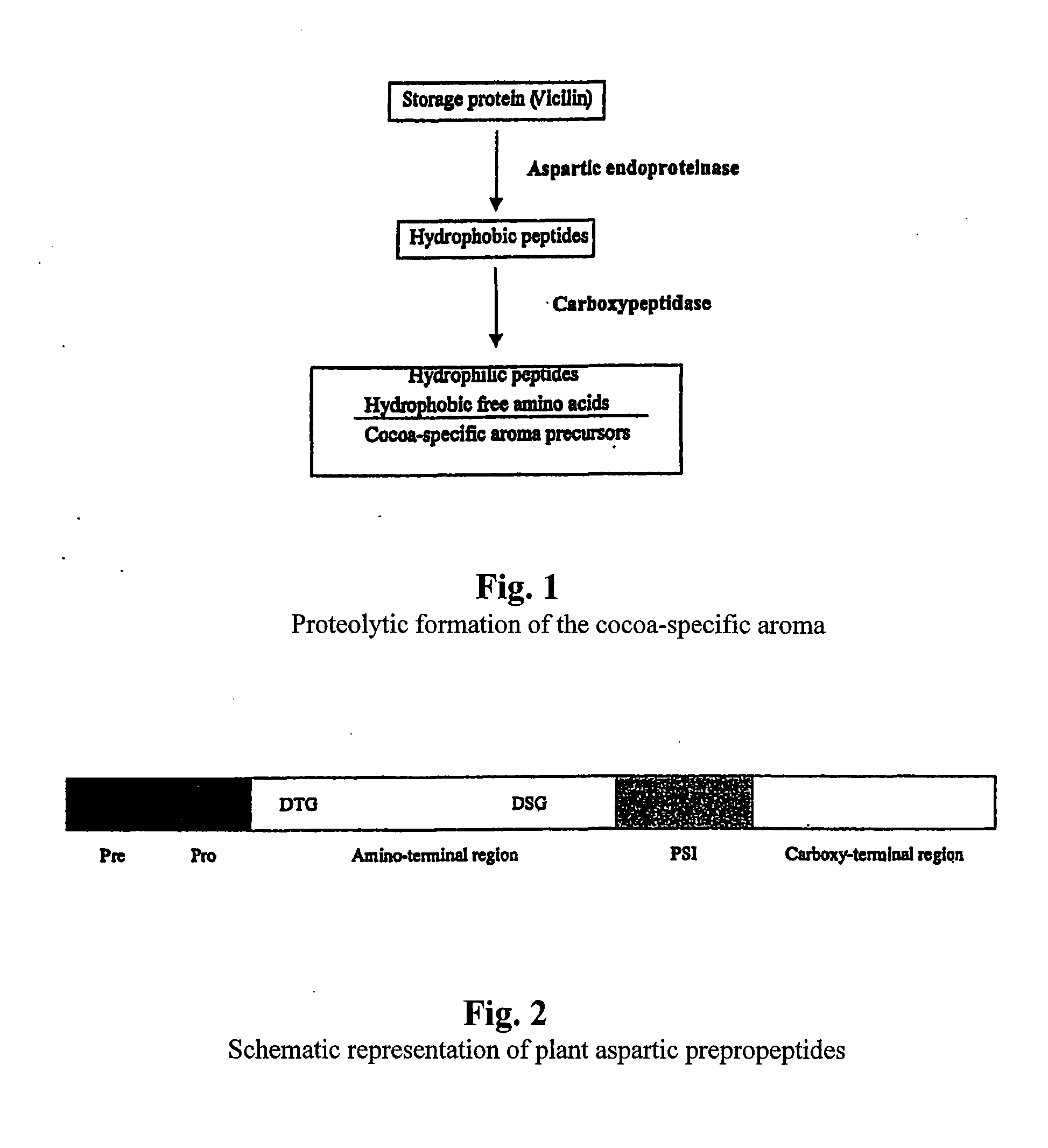
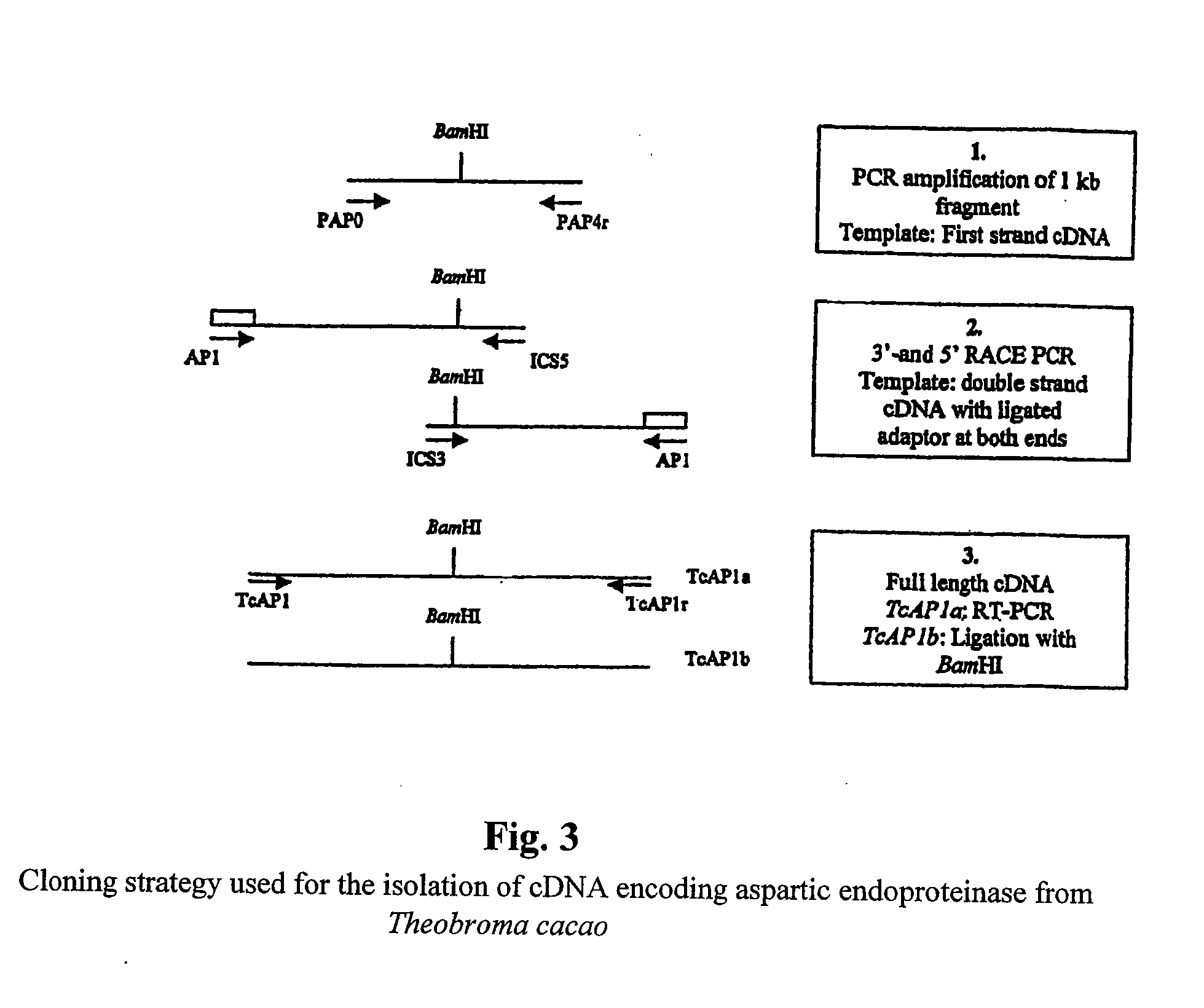
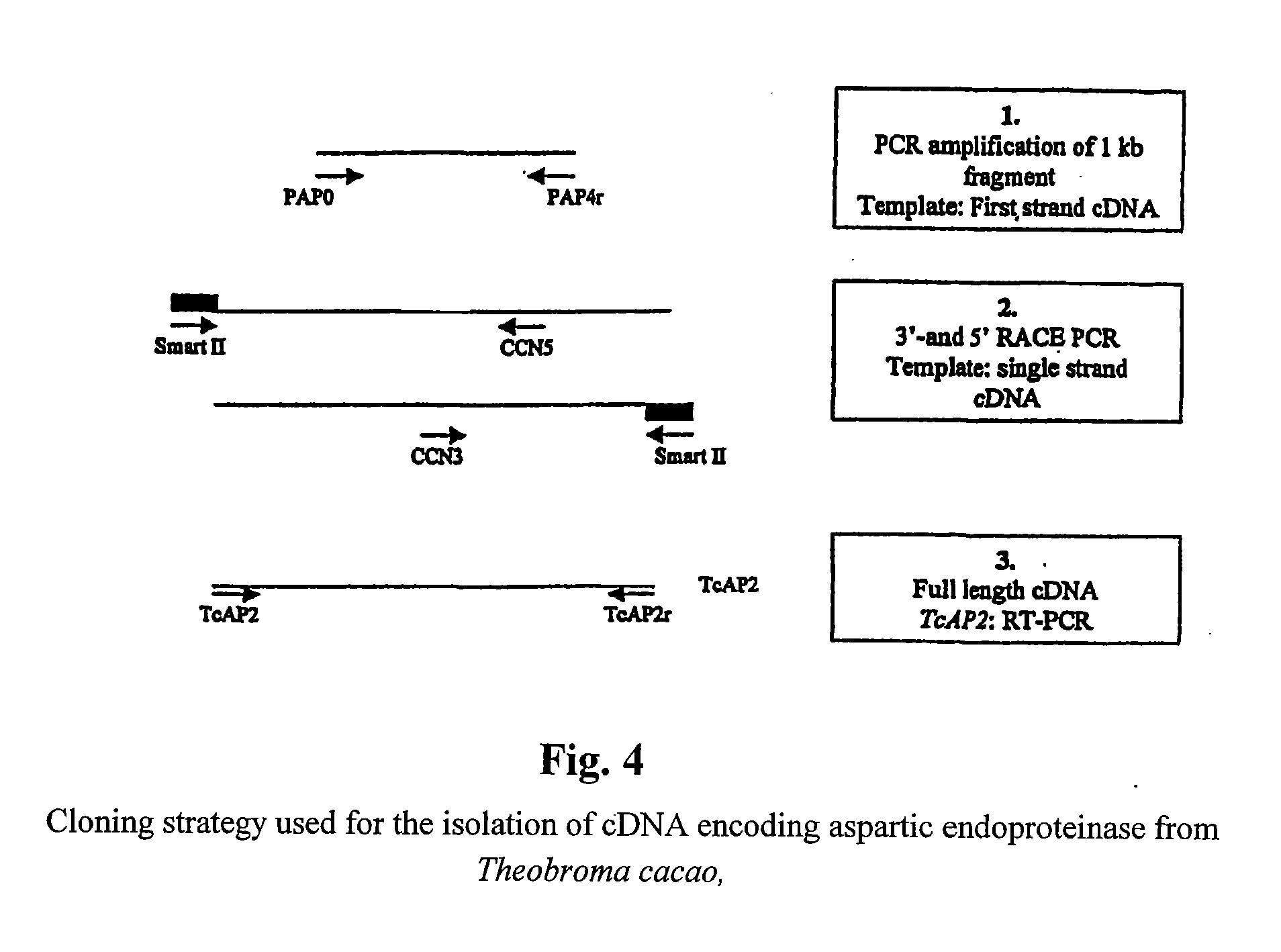
Cacao endoproteinases and production of cocoa flavor from same
a technology of cacao endoproteinase and cocoa flavor, which is applied in the direction of hydrolases, biochemistry apparatus and processes, food preparations, etc., can solve the problem of peptides exhibiting differing sizes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of mRNA
[0087] Two beans were grounded in liquid nitrogen to a fine powder and extraction was directly performed with a lysis buffer containing 100 mM Tris HCl pH8, 1% SDS and 0.1M β-mercaptoethanol. RNA was extracted with one volume phenol / chloroform / isoamylalcohol (25 / 24 / 1) and centrifuged at 8000 rpm for 10 min at 4° C. The aqueous phase was washed three times with chloroform / isoamylalcohol (24 / 1). RNA was precipitated with 0.3M sodium acetate pH 5.2 in two volumes of ethanol. The RNA pellet obtained after centrifugation was resuspended in 100 mM Tris HCl pH 8 and a second precipitation with 2M lithium chloride was performed. The RNA pellet was washed with 70% ethanol and resuspended in DEPC treated water.
example 2
Cloning of Aspartic Proteinase cDNAs
[0088] A search for aspartic proteinase sequences in the GenBank database led to the identification of several plant sequences. A multiple alignment of these sequences revealed the presence of conserved regions, which have been used to design two degenerate oligonucleotides:
[0089] A sense primer, pAP0 (SEQ ID NO:7) (5′-GAYACNGGNAGYTCYAAYYTVTGG) has been synthesized according to the sequence Asp-Thr-Gly-Ser-Ser-Asn-Leu-Trp (SEQ ID NO:17), which contains an active site (Asp-Thr-Gly) of the protein.
[0090] An antisense primer pAP4r (5′-CCATMAANACRTCNCCMARRATCC) (SEQ ID NO:8) has been synthesized according to the sequence Trp-Ile-Leu-Gly-Asp-Val-Phe (SEQ ID NO:18), located in the C-terminal part of the protein.
[0091] Total RNA as prepared in example 1 was used to synthesize first strand cDNA with the SMART PCR cDNA Synthesis Kit (Clontech, USA). Synthesis has been performed exactly as described in the kit instructions using 1 μg of total RNA and th...
example 3
Sequencing and Analysis of DNA Sequences
[0104] cDNA sequencing has been performed according to standard techniques (Maniatis, A Laboratory Manual, Cold Spring Harbor, 1992). Sequence analysis and comparison were done using DNAStar programme. The sequences are shown under SEQ ID Nos. 1 and 2.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com