Anti-infective therapy
a technology of anti-infective therapy and anti-tumor, which is applied in the field of anti-infective therapy, can solve the problems of not being able to determine the sequence of dnase-encoding nucleic acids, unable to obtain dnase products from shields et al., and being toxic to host cells, etc., and achieves the effect of being easily expressed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
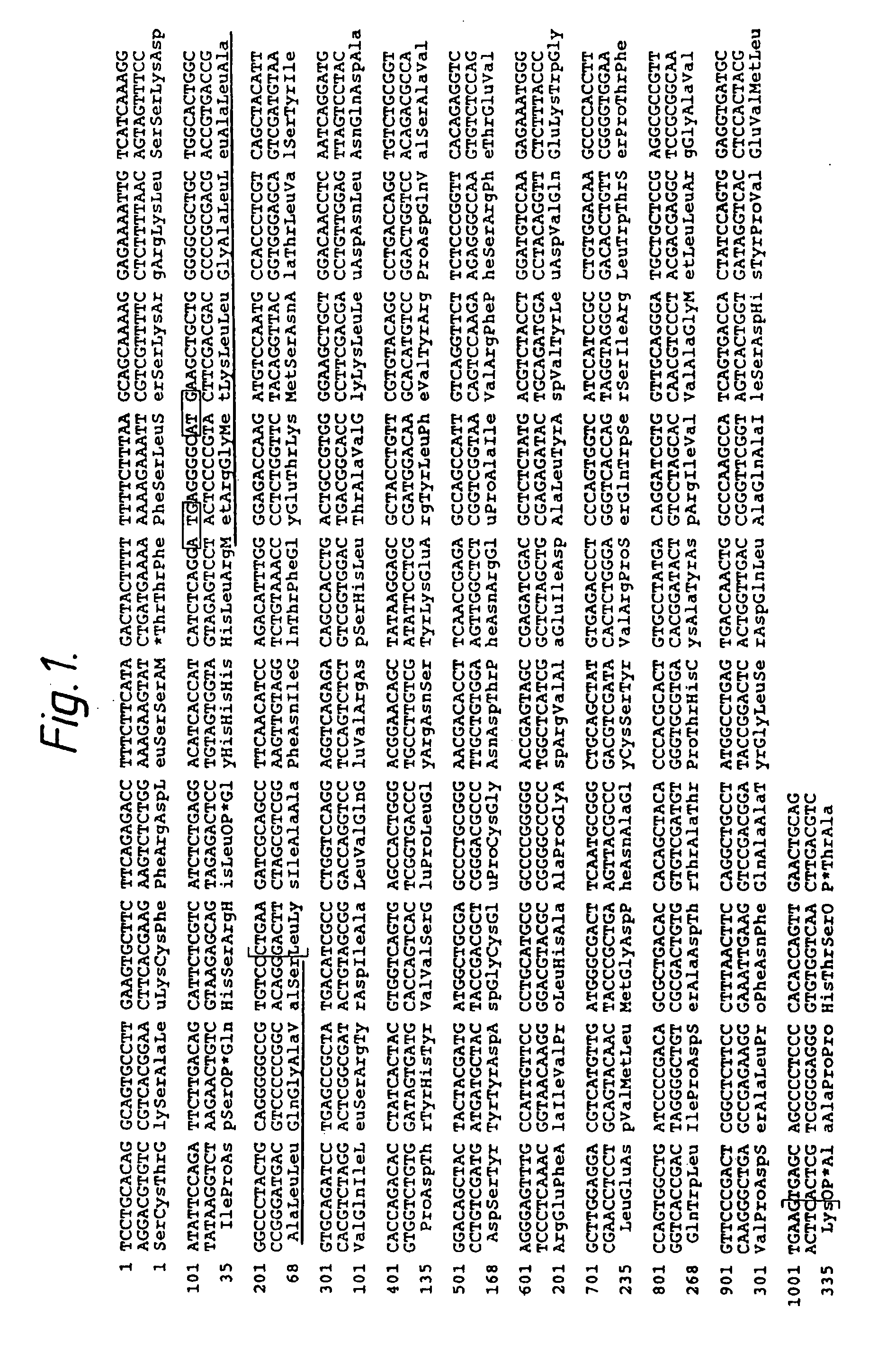
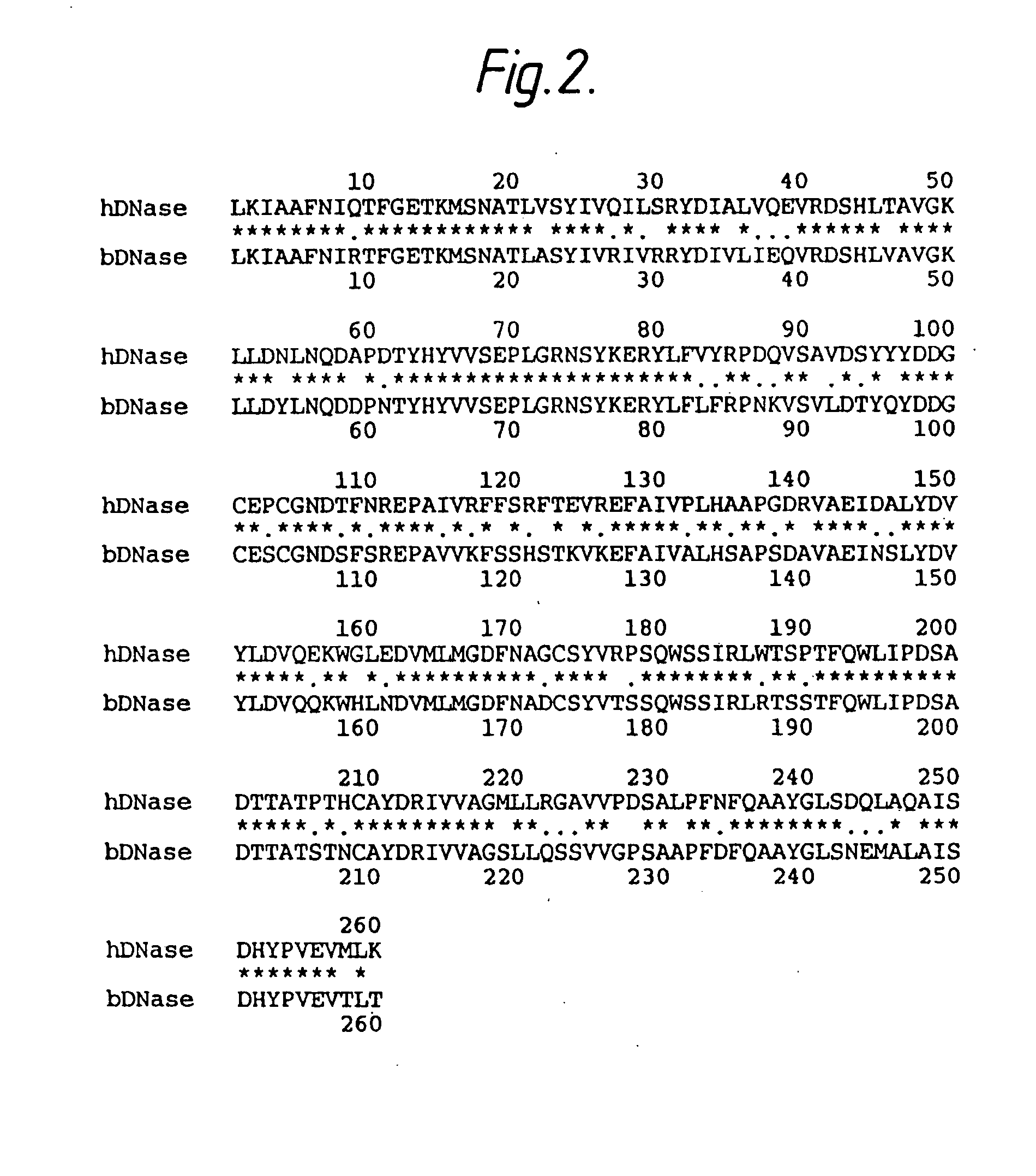
Cloning of Human Pancreatic DNase I
cDNA Library Preparation
[0093] A human pancreatic cDNA library was constructed in λgt10 using polyadenylated mRNA prepared from freshly obtained and liquid N2 frozen human pancreas (Lauffer et al., Nature 318:334 [1985]). Using oligo dT primers and EcoRI-SalI-XhoI-SstII adapters, a cDNA library of 0.9×106 independent isolates of greater than 600 bp was obtained.
Oligonucleotide Probes
[0094] Two long probes were synthesized based on the amino acid sequence of bovine DNase I. Two segments of amino acid sequence which exhibited low redundancy were selected and mammalian codon usage tables were employed.
Probe 1:5′GTG-CTG-GAC-ACC-TAC-CAG-TAT-GAT-GAT-GGC-TGT-GAG-TCC-TGT-GGC-AAT-GAC 3′ (51 mer correspondingto the amino sequenceVal-Leu-Asp-Thr-Tyr-Gln-Tyr-Asp-Asp-Gly-Cys-Glu-Ser-Cys-Gly-Asn-Asp)Probe 2:5′TAT-GAC-GTC-TAC-CTG-GAC-GTG-CAG-CAG-AAG-TGG-CAT-CTG-AAT-GAT-GTG-ATG-CTG-ATG-GGC-GAC-TTC-AAC-GC3′ (71 mer corresponding to the amino acidsequence Ty...
example 2
Assays for DNase Activity
[0102] In addition to standard ELISA (see example 4) and RIA, three assays have been developed to detect DNase activity.
[0103] 1. Hydrolysis of 32P-labeled DNA. Radiolabeled 32P-DNA was prepared using an M13 single-stranded template, a 17mer sequencing primer, 32P-dCTP, non-radioactive DATP, dTTP, and dGTP, and Klenow. Briefly, 1.5λ MgCl2 (35 mM), 1.5λ 10× restriction buffer (70 mM Tris-HCl, pH 7.6; 35 mM dithiothreitol; 1 mM EDTA), and 6λ H2O were mixed with 1 template (approximately 1 μg) and 1.5λ Palmer (0.5 μM) and heated to 55° C. for 7 min. Nucleotide mix was prepared by taking 40λ32P-dCTP (sp. act.3000 Ci / mmol; 400 μCi) plus 1λ each of 2 mM stocks of non-radioactive DATP, dTTP, and dGTP to dryness and then reconstituting the nucleotides in 7λ 1× restriction buffer. The nucleotide mix and 1λ Klenow were added to the template-primer mixture and the reaction was incubated at 37° C. After 15 min, 2λ of a non-radioactive deoxynucleotides were added and t...
example 3
Expression of Human DNase I
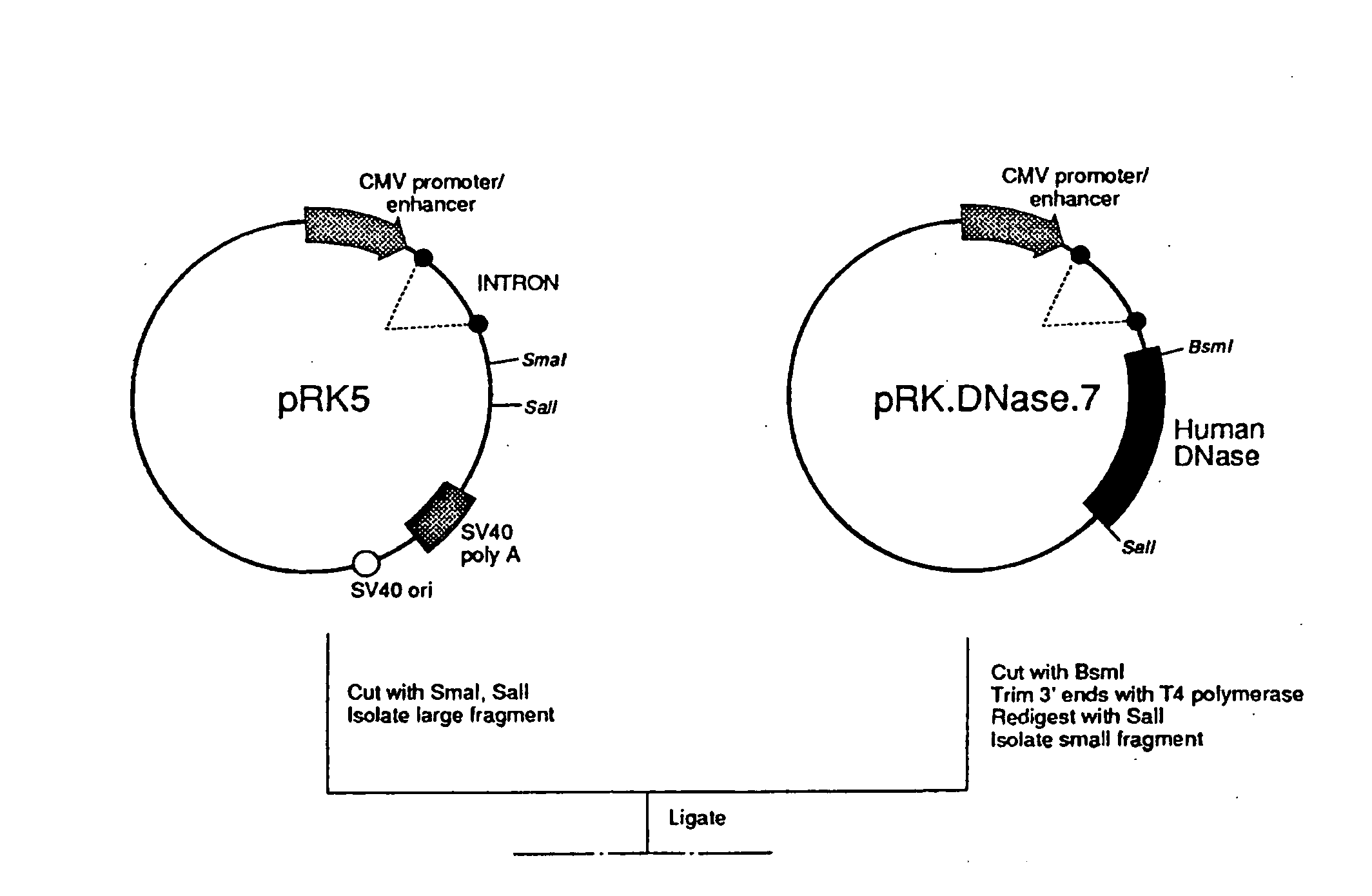
1. pRK.DNase.7
[0108] Plasmid pRK.DNase.7 was constructed from clone 18-1, described above, as follows:
[0109] The plasmid pRK5 was digested with EcoRI, dephosphorylated, and fragment 1 comprising the bulk of the plasmid was isolated. pRK5 is described in Suva et al., Science 237:896 (1987); U.S. Ser. No. 97,472, filed Sep. 11, 1987; and EP Publ. 307,247, published 15 Mar. 1989, where the pCIS2.8c28D starting plasmid is described in EP 278,776 published Aug. 17, 1988 based on U.S. Ser. Nos. 07 / 071,674 and 06 / 907,297. The λ DNase clone 18-1 was digested with EcoRI and the insert (fragment 2) was isolated. Fragment 1 and fragment 2 were ligated and the ligation mixture transformed into E. coli strain 294. The transformed culture was plated on ampicillin media plates and resistant colonies selected. Plasmid DNA was prepared from transformants and checked by restriction analysis for the presence of the correct fragment.
[0110] pRK.DNase.7 was transfected int...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com