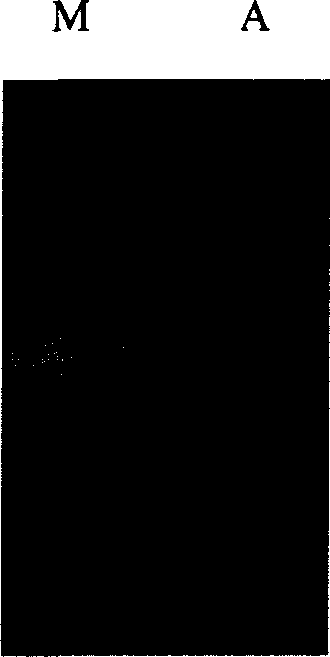Gene for coding, 1,3-propylene glycol reductase in E.aero strain
A technology of propylene glycol and reductase, which is applied in the directions of oxidoreductase, genetic engineering, plant genetic improvement, etc., can solve problems such as low yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1: CGMCC 0532 bacterial strain genome DNA extraction
[0029]After CGMCC 0532 strain of the present invention is activated, inoculate to 8ml LB liquid culture medium (component concentration is 1% (W / V) peptone, 0.5% (W / V) yeast extract, 1% (W / V) ) NaCl) at 30°C overnight. The above-mentioned liquid seeds cultivated overnight are inoculated in 200 ml LB liquid medium according to the inoculum amount of 3-5%, and cultured at 30° C. to the mid-logarithmic growth phase. Centrifuge the above bacteria solution at 10000rpm at 4°C for 10-15min, and discard the supernatant. The collected bacteria were suspended in 2 ml of sterile pure water, centrifuged at 5000 rpm for 10 min at 4°C, discarded the supernatant, and repeated this operation twice. Add 400 μl sterile pure water to the collected bacteria, shake and suspend on a shaker, add 100 μl lysozyme (10 mg / ml) to make the final concentration reach 2 mg / ml, and keep the temperature in a super constant temperature w...
Embodiment 2
[0031] Embodiment 2: dhaT gene PCR cloning
[0032] 1. Cloning dhaT gene by PCR method
[0033] 1.1 Design and synthesis of primers
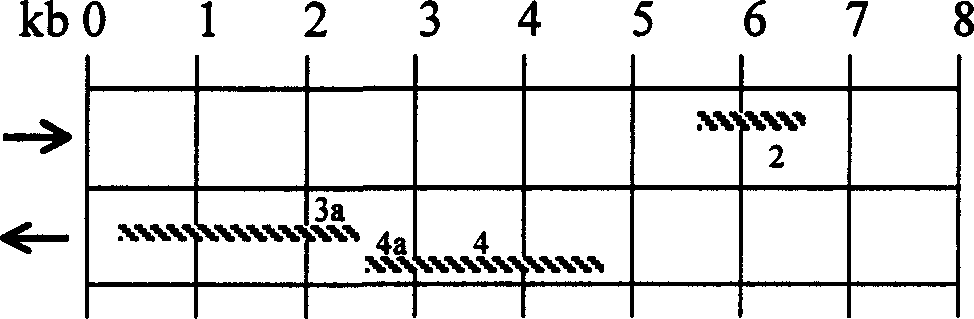
[0034] According to the dhaT gene of the K.pneu bacterial strain (as shown in SEQ ID NO: 3), design and synthesize the oligonucleotide primers used for dhaT gene amplification, and design a Hind III restriction site at the 5' end of the upstream primer , EcoR I restriction site was designed at the 5' end of the downstream primer.
[0035] PCR primers:
[0036] dhaT F 5′- AAG CTT ATG AGC TAT CGT ATG TTT GAT TAT CTG-3' (SEQ ID NO: 4)
[0037] dhaT R 5′- G AAT TC T CAG AAT GCC TGG CGG AAA AT-3' (SEQ ID NO: 5)
[0038] 1.2 PCR cloning dhaT gene
[0039] Use LA Taq TM , pyrobest Taq, EX Taq TM Kits (purchased from Treasure Bioengineering (Dalian) Co., Ltd., product numbers are: DRR002A or DRR002B; DR005A or DR005B; DRR001A or DRR001B or DRR001C), using CGMCC 0532 strain genomic DNA as a template, dhaTF / dhaTR as primers, as PCR.
[0040]...
Embodiment 3
[0071] Example 3: dhaT gene expression
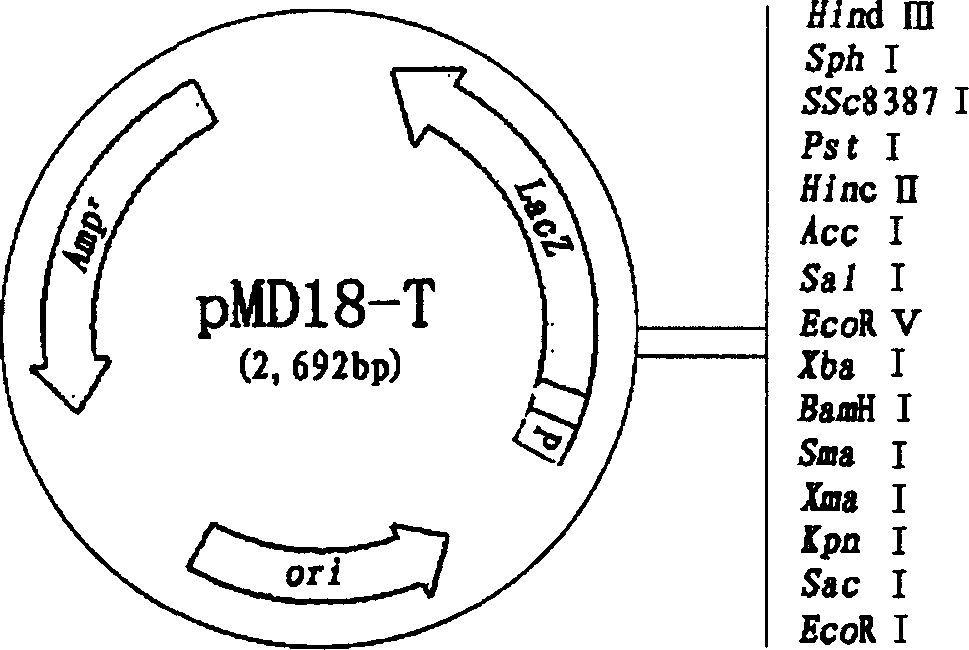
[0072] Take the pMAL-c2X plasmid (purchased from New England Biolabs, product number: E8000S, see the structure Figure 4 ) constructed the fusion gene expression vector pMC58 (see the structure Figure 5 ). Sequencing results showed that the dhaT gene on the pMC58 expression vector was a full-length dhaT gene.
[0073] In 1 liter of LB medium, insert the culture of Escherichia coli JM109 containing pMC58 according to the inoculation amount of 1%, add IPTG to make the final concentration reach 0.4-0.8mM, the fusion protein expressed by the fusion gene composed of malE gene and dhaT gene Show that fusion protein molecular weight is 85KDa through SDS-PAGE electrophoresis, expression electrophoresis result (see Image 6 , band B). Image 6 Middle A: induced cells, B: purified fusion protein, C: purified dhaT protein, D: uninduced cells, Mh (kDa): 200, 160, 97, 4, 66, 45, ML (kDa): 97, 4, 66, 45, 31, 21.5, 14.5.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com