Method of producing adenosylmethionine
An adenosylmethionine, nucleotide sequence technology, applied in the fields of molecular biology and metabolic engineering, can solve the problems of low fermentation yield, troublesome purification, increase fermentation cost, etc., and achieve high biotransformation rate and bioaccumulation. The effect of high, low cost of culture medium
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
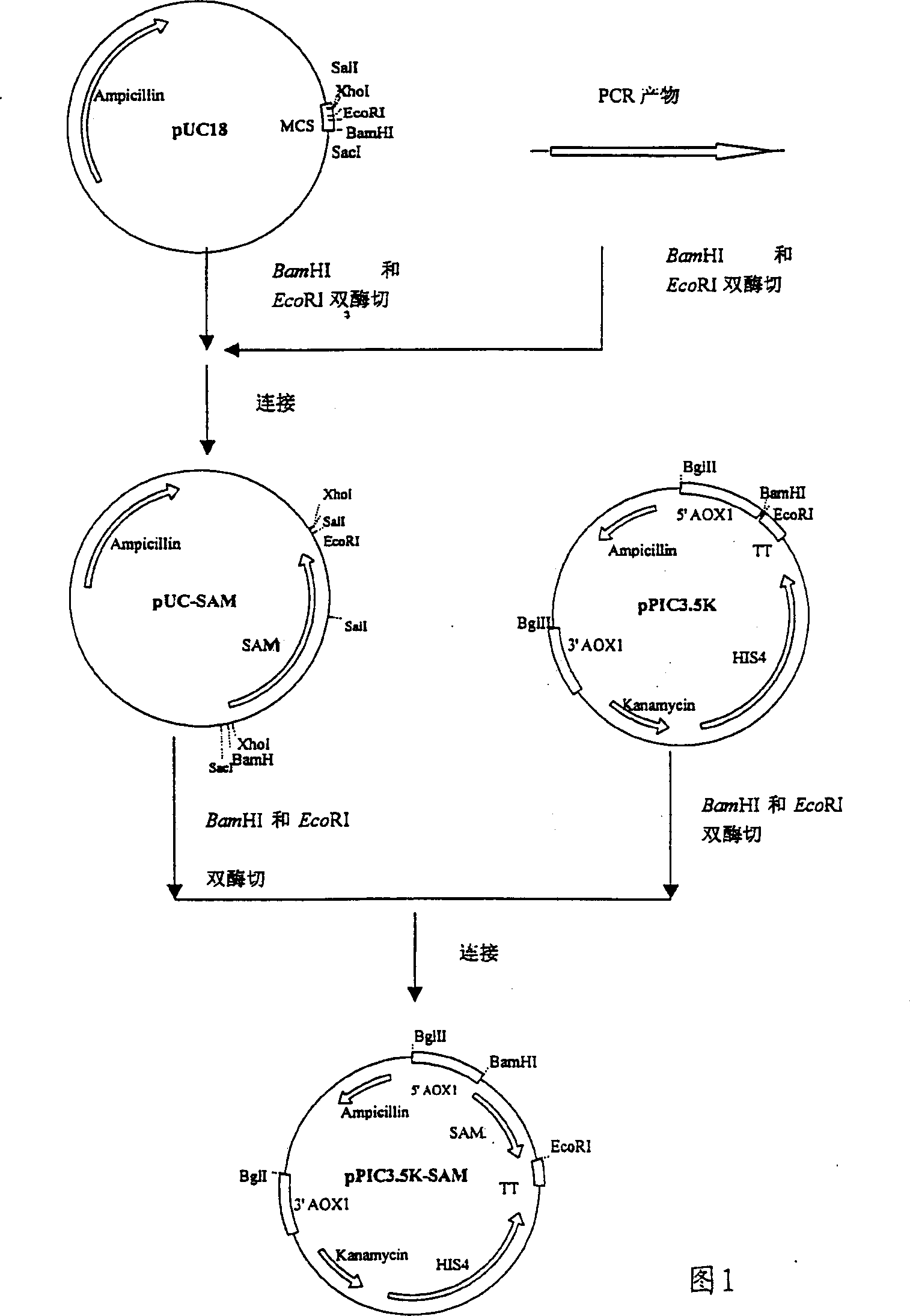
[0024] Construction of recombinant expression SAM synthetase plasmid pPIC3.5K-SAM (see Figure 1)
[0025] 1. Extraction of Saccharomyces cerevisiae chromosomal DNA
[0026] Saccharomyces cerevisiae chromosomal DNA was extracted according to the method of Ausubel et al. (Ausubel, F.M. et al, 1992, Short protocols in Molecular Biology, 2nd edition, P13-43). Inoculate Saccharomyces cerevisiae strain YPH499 in 5ml YPD medium (10 g / L yeast extract, 20 g / L peptone, 20 g / L glucose), and culture at 28-30°C for 16-24 hours. Centrifuge at 5000rpm / min for 5min to collect the bacteria. After washing the bacterial pellet with 0.5ml cold sterile water, resuspend in 0.5ml sorbitol solution (0.9M sorbitol, 0.1M Tris-Cl, ph8.0, 0.1MEDTA), add 50ul of 0.3mg / ml Zymolase and 50ul of 0.28M 2-mercaptoethanol, mix well and react at 37°C for 1 hour. Centrifuge at 5000rpm for 5min, collect the thalli, wash once with 0.5ml Tris / EDTA solution (50mM Tris-HCl (pH8.0) 20ml) cold sterile water and 20ml 1...
Embodiment 2
[0038] Example 2 Construction of recombinant cells expressing exogenous SAM synthetase
[0039] 1. Preparation of electrotransformed Pichia pastoris cells
[0040] Inoculate Pichia pastoris strain GS115 in 500ml YPD medium (1 g / L yeast extract, 2 g / L peptone, 2 g / L glucose), and cultivate to OD at 28-30°C 600 1.3-1.5, centrifuged at 5000rpm / min for 5min, the cell pellet was washed once with 100ml frozen sterile water, 20ml frozen sterile water and 20ml1mol / L frozen sorbitol respectively, and centrifuged at 5000rpm / min for 5min after each wash. Collect the cells, and finally suspend the centrifuged cells with 200 μl of 1M frozen sorbitol to obtain electroshock-competent cells.
[0041] 2. Electrotransformation of yeast cells with recombinant expression plasmid pPIC3.5K-SAM
[0042] About 10 μg of the constructed recombinant expression plasmid pPIC3.5K-SAM was digested and linearized with SalI, and the linear DNA was recovered by ethanol precipitation and dissolved in 10 μl st...
Embodiment 3
[0045] Example 3 A small amount of rapid extraction of intracellular SAM
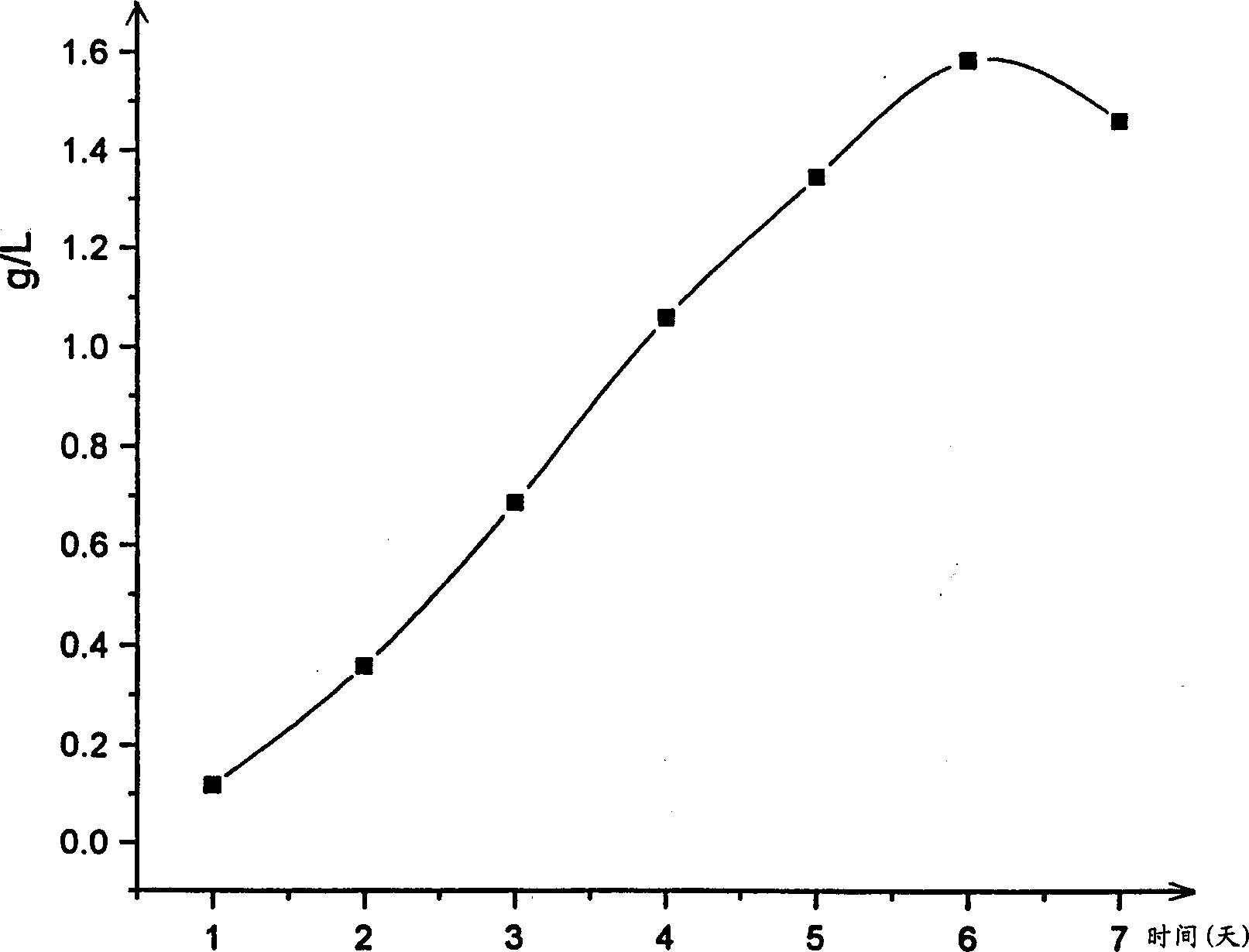
[0046] The fermentation broth with known cell concentration was centrifuged to collect the bacteria, adding 10% trichloroacetic acid equal to the volume of the original fermentation broth to extract at 4°C for more than one hour, centrifuged at 12,000g for 10 minutes, and the supernatant was filtered through a 0.2μm filter membrane After filtration, 20 microliters were taken from it and loaded into HPLC for quantification of SAM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com