Method for simultaneously enhancing and inhibiting multiple key genes in synthesis of saccharomyces cerevisiae 7-dehydrocholesterol
A technology of dehydrocholesterol and Saccharomyces cerevisiae, applied in the direction of microorganism-based methods, botany equipment and methods, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Construction of Saccharomyces cerevisiae producing 7-DHC and related genes in the enhanced pathway
[0046] (a) Using the S288C genome of Saccharomyces cerevisiae as a template, use primers ADH-F and ADH-R to amplify the gene fragment ADH2, use primers IDI-F and IDI-R to amplify the gene fragment IDI1, and use primers E1-F , E1-R amplified to obtain gene fragment ERG1, using primers E11-F, E11-R to amplify to obtain gene fragment ERG11, using primers E24-F, E24-R to amplify to obtain gene fragment ERG24, using primers E25-F, E25 -R to amplify the gene fragment ERG25, use primers E26-F and E26-R to amplify the gene fragment ERG26, use primers E27-F and E27-R to amplify the gene fragment ERG27, use primers GAL1-F, GAL1-R The promoter gene fragment gal1p was amplified, and the promoter gene fragment gal7p was amplified by using primers GAL7-F and GAL7-R. Through gene synthesis, fragments dhcr24 and thmg1 were obtained. Using primers 208UP-F and 208UP-R to ampli...
Embodiment 2
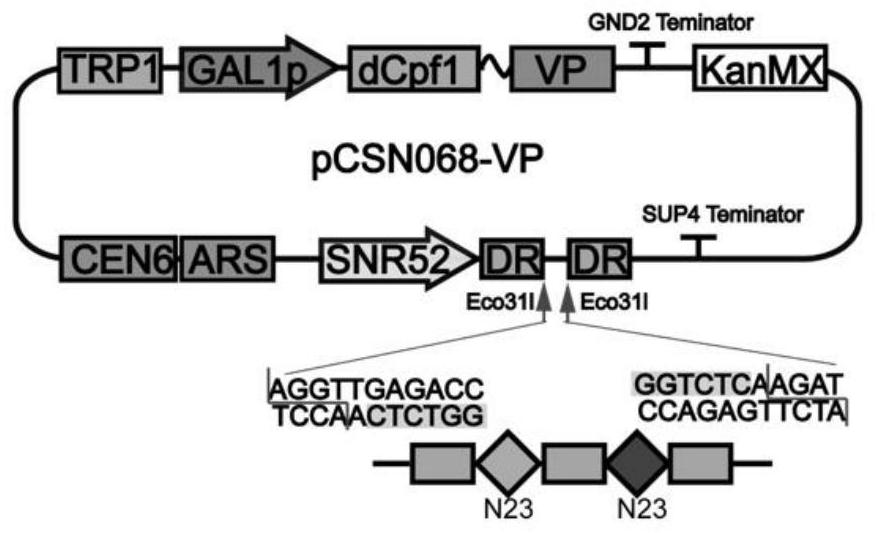
[0101] Construction and application of embodiment 2dCpf1-VP activation system
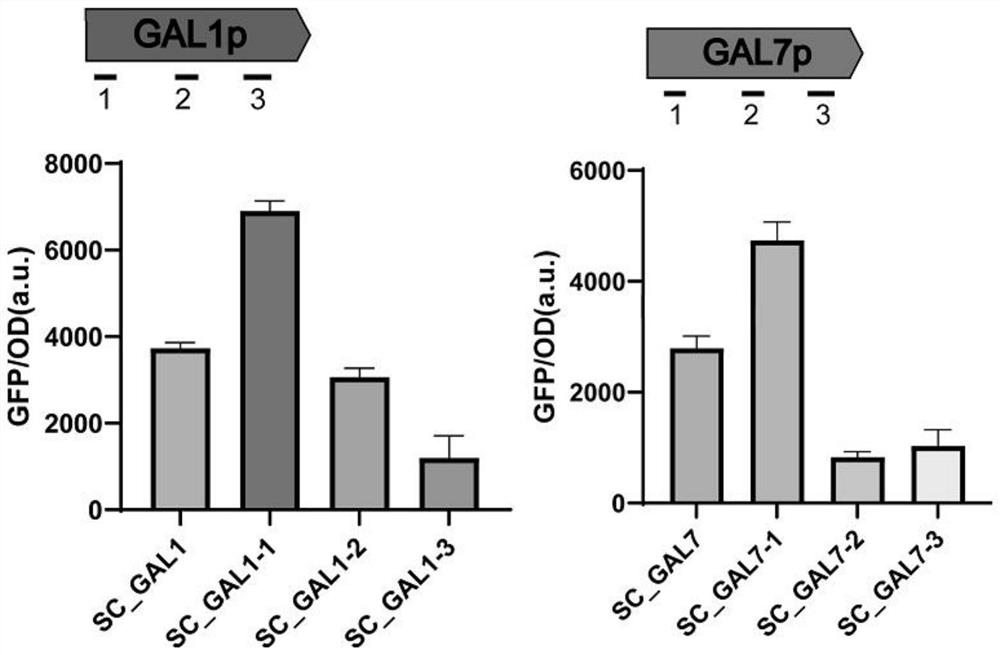
[0102] (a) Using the plasmid pCSN068 purchased from Addgene as a template, Cpf1 was mutated into dCpf1 using primers dCpf1Tu-F and dCpf1Tu-R. The activation domain VP was gene synthesized, and the activation domain VP was connected with pCSN068 containing dCpf1 using seamless cloning to obtain the plasmid pCSN068-VP. Using the plasmid pML104 purchased from Addgene as a template, primers 52p-F and 52p-R were used to amplify the promoter gene fragment SNR52p. The fragment gal1p23-1 was synthesized directly by overlapping primers gal1p23-F1 and gal1p23-R1, and the fragment gal7p23-1 was synthesized directly by overlapping primers gal7p23-F1 and gal7p23-R1. The fragment gal1p23-2 was synthesized directly by overlapping primers gal1p23-F2 and gal1p23-R2, and the fragment gal7p23-2 was synthesized directly by overlapping primers gal7p23-F2 and gal7p23-R2. The fragment gal1p23-3 was synthesized directly...
Embodiment 3
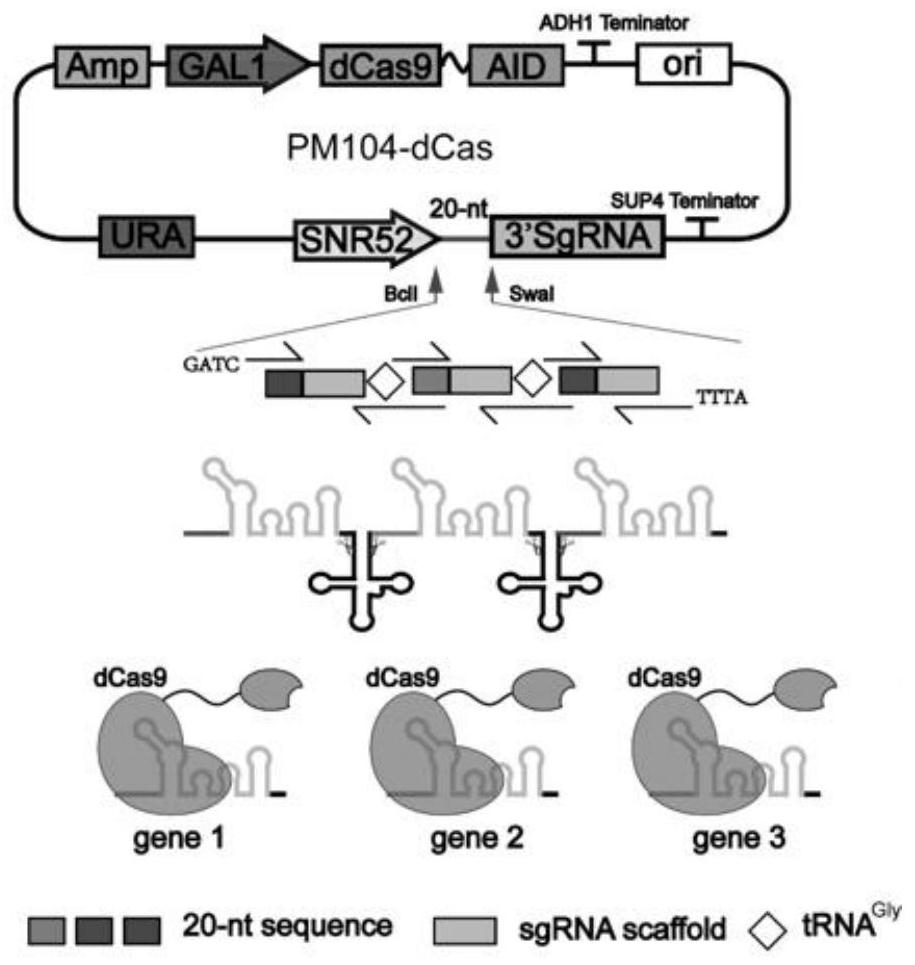
[0145] Construction and application of embodiment 3dCas9-RD suppression system
[0146] (a) Using the plasmid pML104 purchased from Addgene as a template, Cas9 was mutated into dCas9 using primers dCas-F1, dCas-R1, dCas-F2, and dCas-R2. Gene synthesis inhibitory domain RD, use seamless cloning to connect activation domain RD and pML104 to obtain plasmid pML104-RD, use the above constructed plasmid as a template, and use CIT-F and CIT-R as primer loop P to obtain plasmid pML104 -RD-CIT, using MLS1-F and MLS1-R as primer loop P to obtain plasmid pML104-RD-MLS, using ERG6-F and ERG6-R as primer loop P to obtain plasmid pML104-RD-ERG6. The fragments URA-dCas9-RD-CIT, URA-dCas9-RD-MLS, and URA-dCas9-RD-ERG6 were amplified by using URA-104-F and URA-104-R as primers, respectively. Using the plasmid pFA6a-TRP1-PGAL1-GFP purchased from Addgene as a template, the fragment pFA6a-TRP1-GFP was amplified using primers pFA6a-F and pFA6a-R, and using the genome of Saccharomyces cerevisiae S...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com