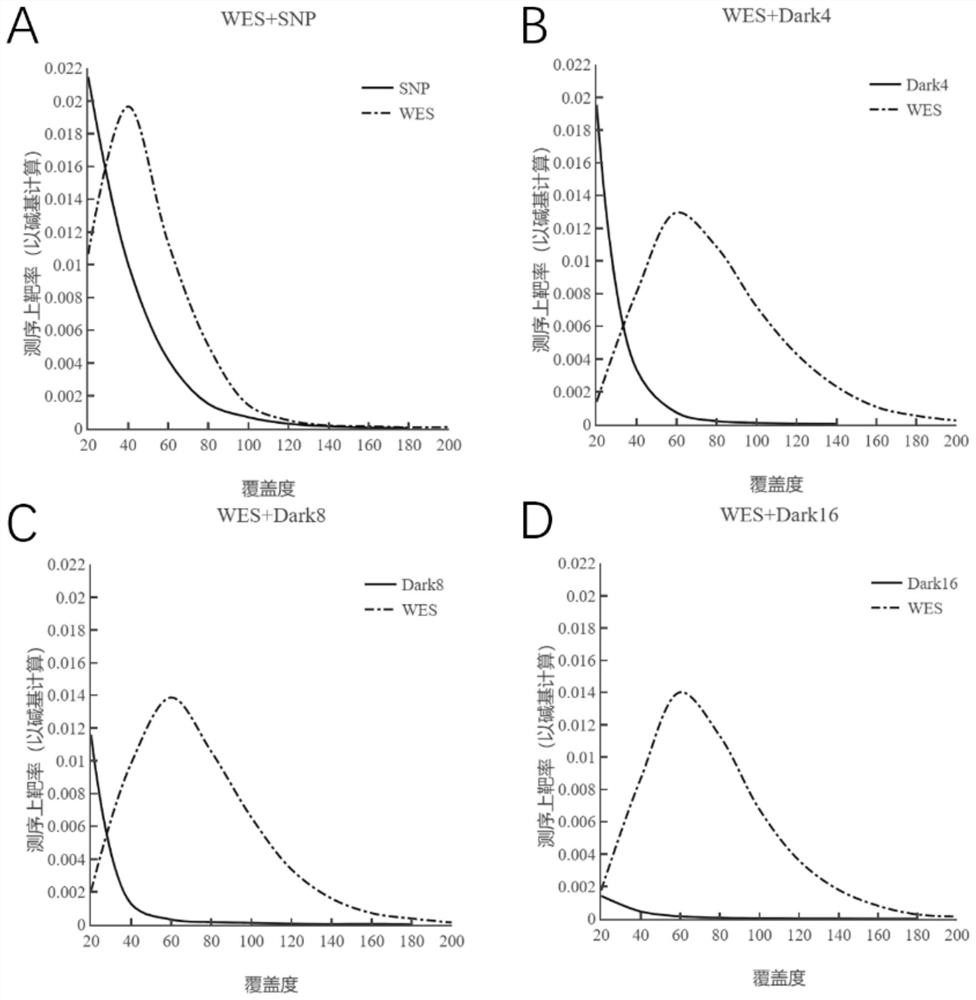
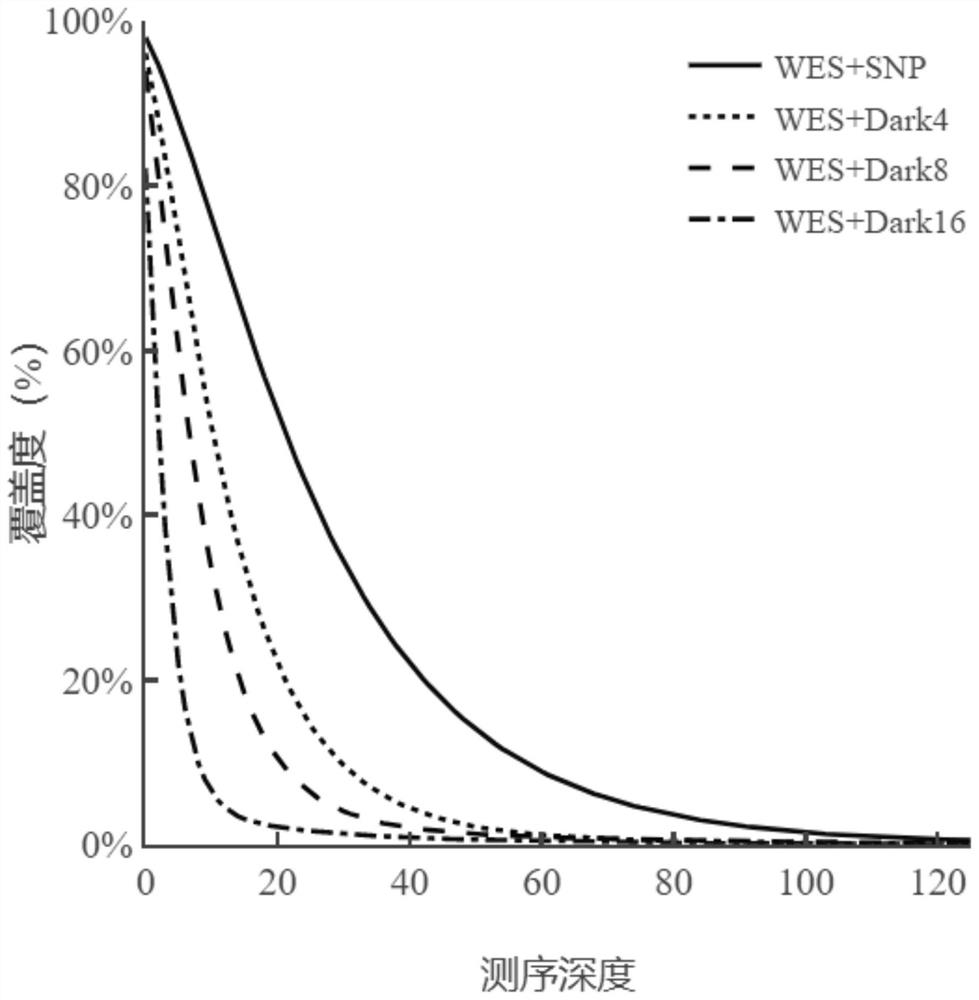
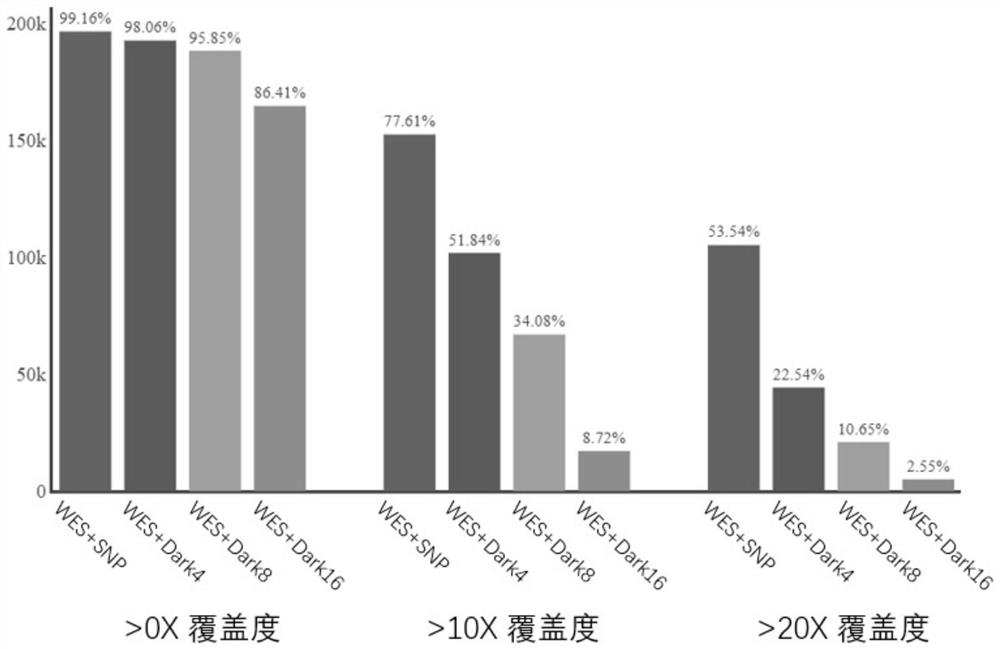
NGS targeted capture method based on dark probe technology and application of NGS targeted capture method in differential depth sequencing
A probe and sequencing technology, applied in the field of high-throughput sequencing library preparation, can solve the problem of not being able to meet the high depth of exon region sequencing at the same time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1. Preparation of all-exon and SNP site liquid capture probes required for WGS
[0043] 1. The human all-exon probe was QuarXeq Human AllExon Probes 3.0 from Shanghai Diying Biotechnology Co., Ltd., product number Y1009A (biotin-labeled), hereinafter referred to as WES. The target region of the WES probe is to use the NCBI Ref-Seq data to cover all the known exon regions.
[0044] 2. The human SNP probe was QuarXeq Human SNP Probes1.0 from Shanghai Diying Biotechnology Co., Ltd., the product number is Y1011A. The probe is a capture probe that covers 200,000 single nucleotide polymorphism sites (SNP) after cross-analysis using multiple Genome-wide association study (GWAS) project databases, and includes standard biotin-labeled probes As with dark probes without biotin labeling (identical probes, the only difference is whether they are labeled with Botin or not), probes labeled with Biotin are hereinafter referred to as SNP probes.
Embodiment 2
[0045] Example 2. Concentration adjustment and ratio of WES probe
[0046] The two types of probe groups in Example 1 were mixed according to different concentrations and ratios. Finally, the probes required for WES were prepared (WES probes were at a fixed concentration, the concentration of SNP probes and the ratio of biotin labeling were adjusted, the final product, WES probes used 300ng, SNPs (including probes with and without biotin labeling) The total amount is 100ng), the operation is as follows:
[0047] For the optimization of the dark probe competition binding scheme for the WES+SNP part, take 3 μL of the biotin-labeled SNP probe stock solution (50ng / μL), add 3 μL of the SNP dark probe without biotin labeling, and mix well. Named SNP-Dark2x, take 3 μL of SNP-Dark2, add 3 μL of SNP dark probe without biotin label, and name it SNP-Dark4x after mixing. Take 3 μL of SNP-Dark4, add 3 μL of SNP dark probe without biotin label, and name it SNP-Dark8x after mixing. Take 3...
Embodiment 3
[0048] Example 3. Liquid phase hybridization capture and sequencing of binary combined probes (WES+SNP, WES+SNP-Dark4x, WES+SNP-Dark8x, WES+SNP-Dark16x)
[0049] 1. Take 200ng NA12878 DNA human gene standard (a human genomic DNA standard, Coriell Institute) for library construction and probe test optimization. The kit used for library construction is QuarPrep Ultra DNALibrary Kit, item number L1001A, Shanghai Diying Biotechnology Co., Ltd. company.
[0050] 2. Start the Covaris S220 system (an ultrasonic DNA fragmenter): make sure that the fresh deionized water exceeds the level 12 on the Covaris tank; pre-cool and degas for half an hour. Add 200ng NA12878 DNA human gene standard, and make up the volume to 50μl with fresh deionized water, then add it to the Covaris micro Tube, stick to the wall and slowly pour out the liquid, being careful not to create air bubbles at the bottom of the tube.
[0051] 3. The ultrasonic conditions on Covaris S220 or M220 are set as shown in Tab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com