Preparation and application of high-salt-resistant and high-activity DNase I mutant
A mutant and nucleic acid construct technology, applied in the fields of genetic engineering and enzyme engineering, can solve problems such as low activity and inactivation, and achieve the effects of reasonable conformation, improved tolerance, and good tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0118] Preparation of samples to be tested:
[0119] Before assaying the activity, dilute the enzyme solution with an appropriate enzyme diluent to within the detection linear range of 200-500U / mL, and detect the enzyme activity according to the following steps.
[0120] DNase I enzyme activity assay steps:
[0121] Add 2.5mL of assay reaction solution R to a 3cm quartz cuvette, keep it warm at 37°C for 1.5min, add 0.5mL of the sample to be tested, mix quickly, and read the average maximum value per minute within 2min at a wavelength of 260nm. Absorbance change rate (ΔA 260 nm / min). Enzyme activity was calculated according to the following formula. The detection linear range is 200-500U / mL. 3.00mL reaction system containing 83mM sodium acetate buffer (pH 5.0), 4.2mM MgSO 4 , 33.33μg / mL calf thymus DNA and 200-500U / mL DNaseI.
[0122] The enzyme activity calculation formula is:
[0123]
[0124] Among them, df: the dilution factor of the initial enzyme solution; 0.0...
Embodiment approach
[0130] The present invention provides polynucleotides encoding DNase I of the present invention. In certain aspects, the invention relates to expression vectors comprising polynucleotides of the invention. In certain aspects, the invention relates to host cells comprising a polynucleotide of the invention. In certain aspects, the present invention relates to a method of producing DNase I, said method comprising: (a) cultivating a host cell of the invention under conditions suitable for expressing said DNase I; (b) optionally recovering said DNase I .
[0131] Embodiment of the expression vector:
[0132] The invention relates to a recombinant expression vector comprising a polynucleotide encoding the DNase I of the invention, a promoter and a terminator. Various nucleotide and control sequences can be linked together to form a recombinant expression vector that can contain one or more suitable restriction sites to allow insertion or substitution of the DNase I variant-encod...
Embodiment 1
[0144] The design of embodiment 1 wild-type DNase I and mutant DNase I coding gene:
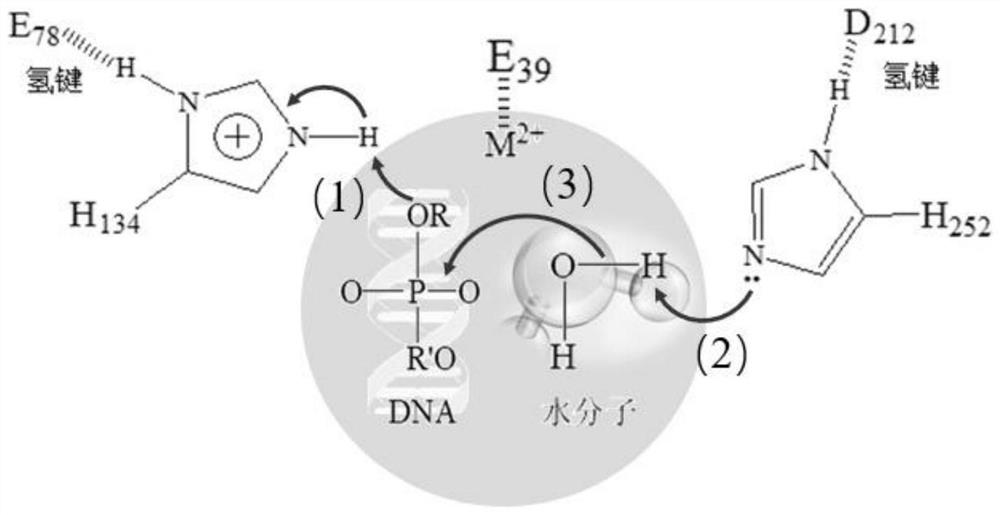
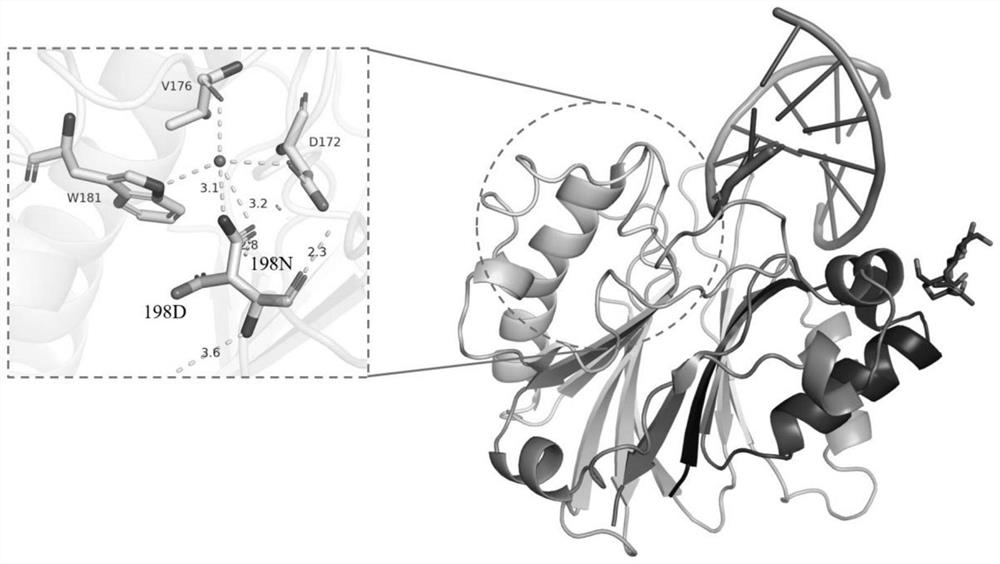
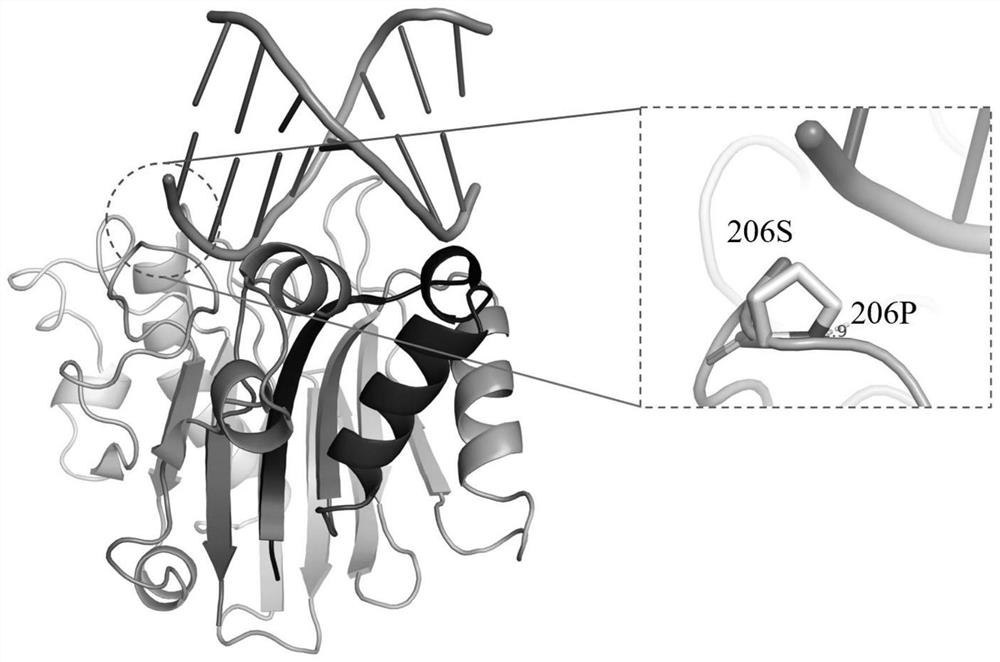
[0145] The key amino acid comparison analysis results of DNase I from different species are shown in Table 2. The 39th E, 78th E, 134th H, 212th D, and 252nd H are active centers, and the 111th R Although it is not the active center, highly conserved amino acids play an important role in catalytic activity; the three-dimensional crystal structure of DNase I derived from bovine pancreas (PDB 1DNK) is shown in figure 2 and image 3 As shown, the ninth R, the 13th E and the 74th N interact with the substrate DNA phosphate backbone; the 198th D and the 206th S are located on the outer surface of the structure and form random coils.
[0146] Table 2 Alignment of key amino acids of DNase I from different species
[0147]
[0148]
[0149] The amino acid sequence shown in SEQ ID NO.2 is reverse-transcribed and optimized according to the codons of Escherichia coli to obtain the nucleotide se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com