Construction method of self-protection DNA enzyme walker and application of self-protection DNA enzyme walker to living cell miRNA detection
A DNA enzyme and construction method technology, applied in the field of construction of self-protected DNA enzyme walker, to achieve high specificity, good stability against degradation, and increase the effect of cleavage rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
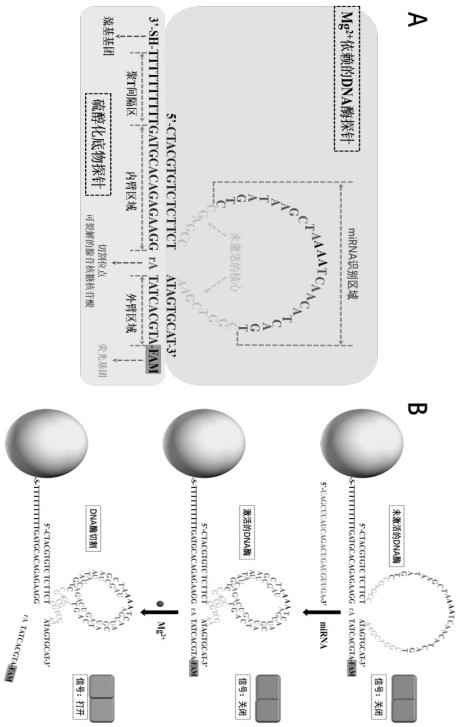
[0039] Example 1 A method for preparing a self-protected DNase walker, comprising the following steps:
[0040] (1) First, gold nanoparticles (AuNPs) with a diameter of 13 nm were prepared by the sodium citrate reduction method. Next, in 19.5 µL of 500 mM acetate buffer (Tris-Ac buffer pH 5.2), 0.5 µL of 10 mM tris(2-carboxyethyl)phosphine was mixed with 30 µL of 100 µM thiolated substrate chain probe, and the Incubate for 1 hour at room temperature to reduce sulfhydryl groups.
[0041] (2) Add 3 µL of 100 µM Mg to the reduced thiolated substrate chain solution 2+DNase-dependent probes, followed by incubation for 3 hours at 37 °C for complete hybridization. The molar ratio of the two nucleic acids is 1:10 (substrate probe: DNase probe). Then, the resulting mixture was added to a clean glass vial containing 1 mL of AuNP solution (10 nM), and the glass vial was incubated with shaking in the dark (200 rpm) for 16 h. After the incubation time has elapsed, add 10.3 µL of 500 mM...
Embodiment 2
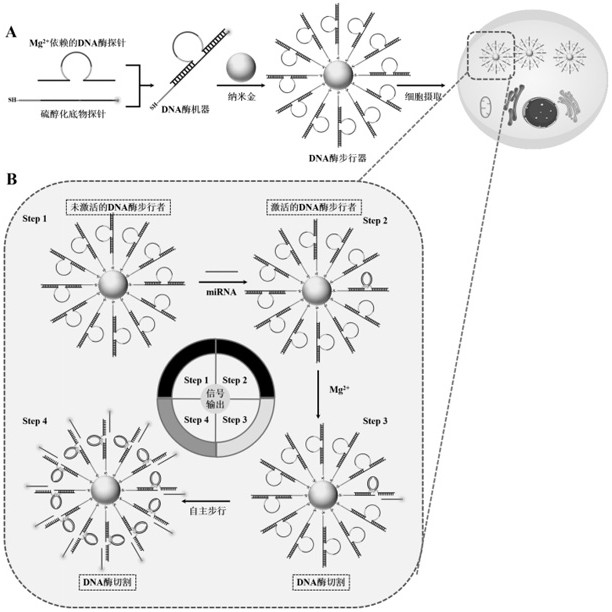
[0047] Example 2 Feasibility analysis of a self-protected DNase walker for in vitro detection of miRNA
[0048] Using the self-protected DNase walker constructed by the method in Example 1, the feasibility analysis was performed with the miRNA-21 target probe.
[0049] The miRNA-21 target probe sequence used is: 5'-TAGCTTATCAGACTGATGTTGA-3'.
[0050] The specific steps of feasibility analysis are as follows:
[0051] First, add Mg to 18 μL Tris-Ac Buffer solution with pH 5.2 2+ Dependent DNase probe (0.1 μM) and thiolated substrate probe (1 μM), then incubated at 37 °C for 3 h to allow Mg 2+ The dependent DNase probe fully hybridizes to the thiolated substrate probe. Next, add 1 μL of 2 μM miRNA-21 target probe to activate the DNase machinery. After incubation at 37 °C for 30 min, add 1 μL of 200 mM Mg 2+ to initiate DNase-catalyzed cleavage. The resulting solution was incubated at 37°C for 8 hours. Other samples, including blanks and controls, were prepared with corres...
Embodiment 3
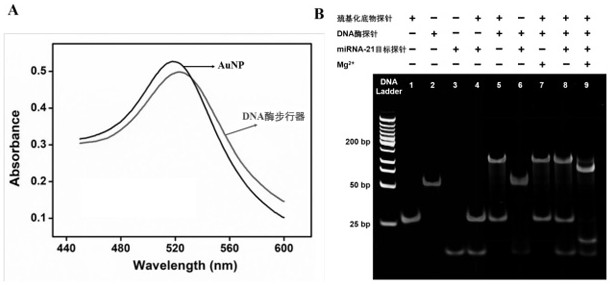
[0054] Example 3 Analysis of the detection ability of the self-protected DNase walker to the target DNA probe
[0055] A self-protected DNase walker was constructed using the method in Example 1, by adding different concentrations of target DNA probes, and then using a fluorescence spectrometer to test its kinetics. Specific steps are as follows:
[0056] Under the condition of 37 ̊C, the time-varying curves of fluorescence signals of different concentrations of miRNA-21 target probe samples and blanks were monitored, and the scanning time was 1 h. The excitation wavelength was 488 nm and the emission wavelength was 520 nm. Specifically, a final concentration of 2 nM of DNase walker and different concentrations of miRNA-21 target probe (final concentrations of 0, 100 pM, 600 pM, 1 nM, 4 nM, 8 nM, 10 nM, 12 nM and 15 nM), and adjust the volume to 190 μL with Tris-Ac buffer (pH = 8.2). After the mixed solution was incubated at 37 ̊C for 30 minutes, 10 μL of Mg 2+ solution (fi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com