Specific sequence for sex determination of snakehead and application
A technology of sex and snakehead, which is applied in the specific sequence and application field of the sex identification of snakehead, can solve the problems of insufficient conservation, low accuracy of gender identification markers, unclear X and Y chromosome conservative sequences, etc., and achieve fish body Less damage, reduced analysis data volume, quality and efficiency improvement effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
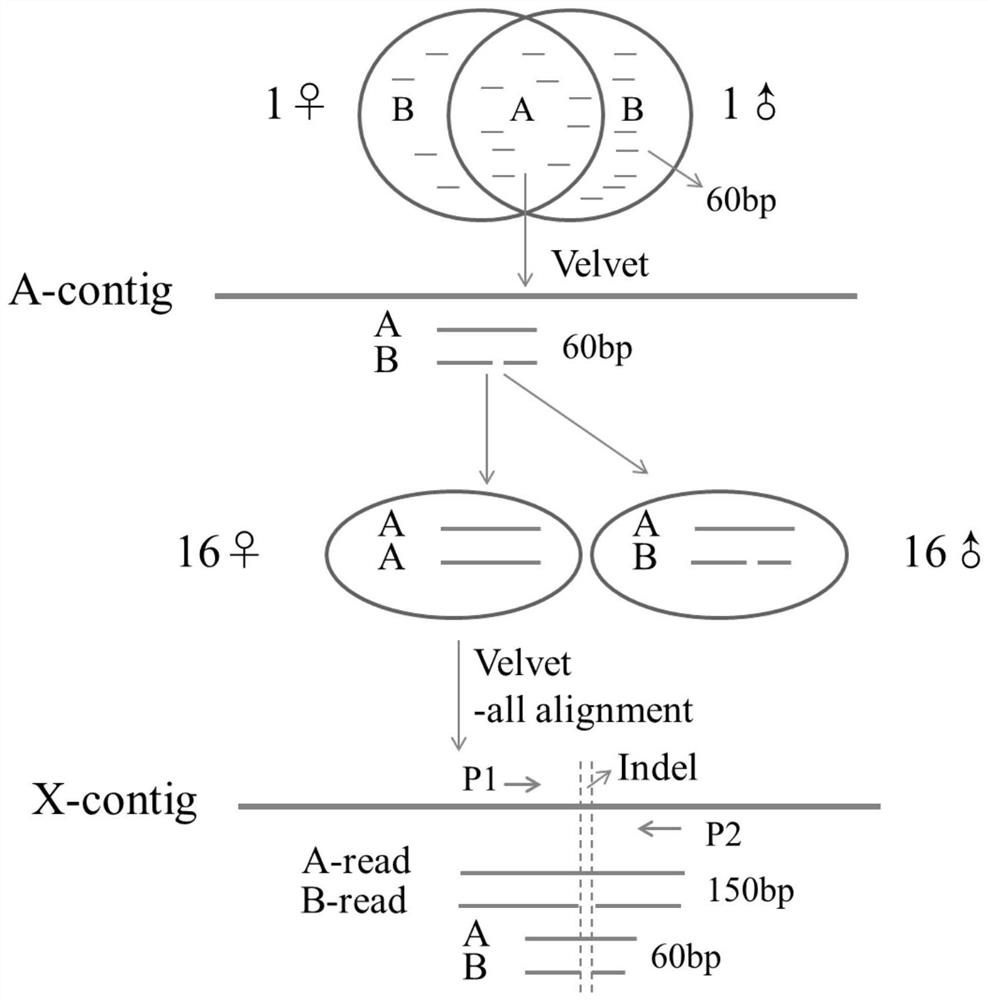
[0026] Acquisition of Indel markers for sex identification of snakehead:
[0027] 1) Select 16 females and 16 males in different waters with sex determined, and perform whole-genome sequencing, and the data volume obtained is about 30G / tail. Select one female and one male sequencing data respectively, and carry out 60bp electronic enzyme digestion treatment starting from six bases of AC, AG, AT, GA, GC and GT. The frequency of various restriction enzyme fragments was obtained after removing redundancy. According to the frequency distribution, the enzyme-cut fragments within the frequency range of the single-copy site were determined and compared to obtain the consensus fragment (A) and the sex-specific fragment (B).
[0028] 2) Consensus fragment (A) was assembled using Velvet software to obtain A-contig. Take A-contig as reference, A and B as query, and use Boutai2 software to compare the default values. In the output result, allelic fragments are obtained by pairing accor...
Embodiment 2
[0034] The application process of the sequence shown in SEQ ID NO.3 in the sex identification of snakehead:
[0035] 1) Design primers for the nucleotide sequence comprising the sequence shown in SEQ ID NO.3:
[0036] CAYSPE-F:GTGTCAATTGTGAGTCCTTGATG
[0037]CAYSPE-R:CCATGCTCTGATCAGTAAATACAC
[0038] 2) Genomic DNA extraction:
[0039] Add 150 μl of 5% chelex100 to a 96-well plate equipped with caudal fins, digest at 58°C for 1 hour, boil at 100°C for 8 minutes, centrifuge at 4000 rpm for 5 minutes, and take 1 μl of the supernatant as a PCR reaction template.
[0040] 3) PCR amplification:
[0041] Configure the reaction system in a 96-well plate, the reaction system is 10×Buffer (Mg 2+ plus) 1μl; LC Green 1μl; dNTP (10mM each) 0.2uL; CAYSPE-F (10mM) 0.2uL; CAYSPE-R (10mM) 0.2uL; Taq (5U / uL) 0.1uL; template DNA 1μl, and finally add ddH 2 0 to 10 μl. The PCR reaction conditions were pre-denaturation at 94°C for 4 min; denaturation at 94°C for 30 s, annealing at 55°C for 3...
Embodiment 3
[0053] The application process of the sequence shown in SEQ ID NO.2 in the sex identification of snakehead:
[0054] 1) Collect 36 male and female snakeheads of known gender, and 12 extra males. Collect fin ray tissue samples and store them in a 96-well plate for cryopreservation. Genomic DNA is extracted by chelex100 boiling method.
[0055] 2) Utilize the method in Example 2 to carry out PCR amplification on the above-mentioned snakehead DNA sample.
[0056] 3) PCR amplification products were subjected to HRM typing.
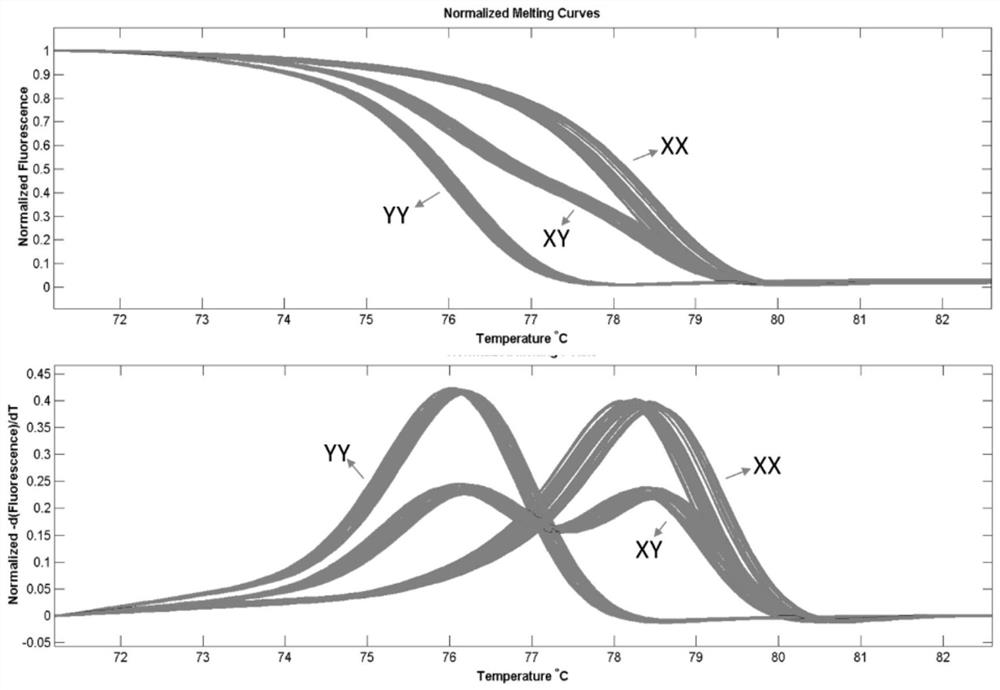
[0057] 4) The HRM typing results show that the identification results using the Indel markers provided by the present invention are completely consistent with the known results, and the identification accuracy rate reaches 100%.
[0058] 5) Test results such as figure 2 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com