Molecular marker for detecting wheat backbone germplasm weekly 8425B specific chromosome segment and application of molecular marker
A technology of chromosome fragments and molecular markers, which is applied in the field of molecular markers for the detection of 8425B specific chromosome fragments of wheat backbone germplasm, can solve the problems of lack of specific detection molecular markers, and achieve the effect of improving detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
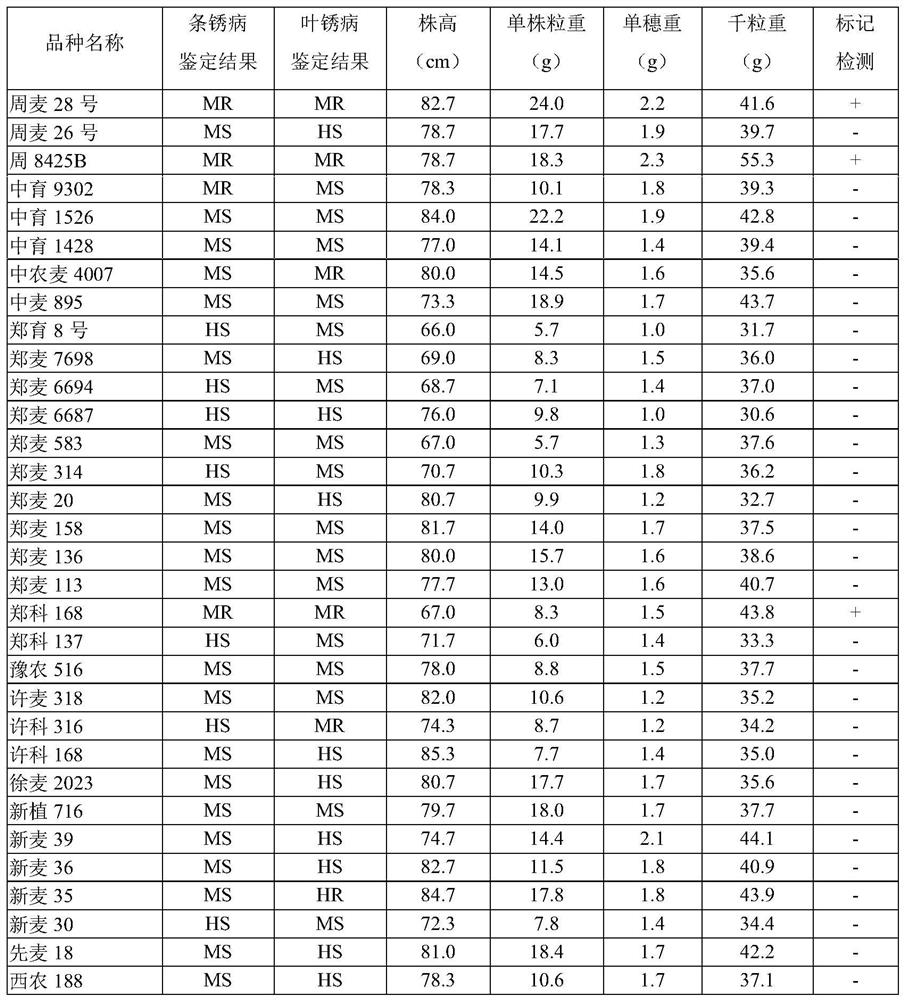
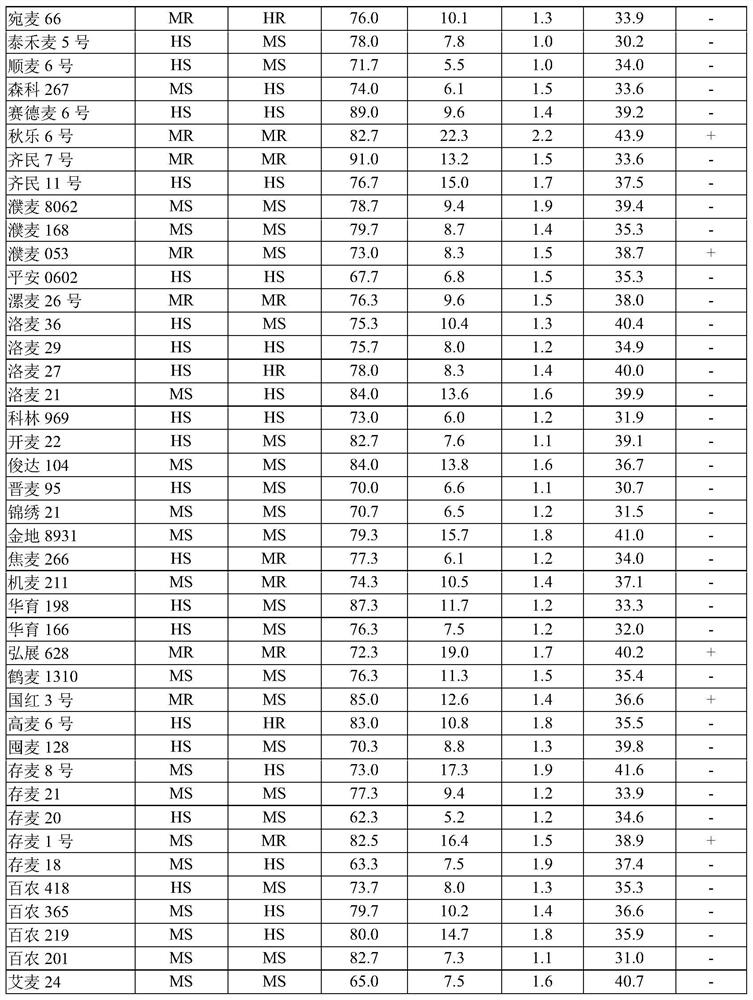
[0034] Specific detection of germplasm resources: The parents of wheat backbone germplasm Zhou 8425B, Guangmai 74, Lianfeng 1, Shanqianmai and Anong 7959, collected other well-known 1BL / 1RS wheat germplasms at home and abroad, Aimengniu, Luomengniu Fulin No. 10, Lovelin No. 13, Lankao 906, Niuzhut, Aurora, Caucasus Mai, Bainong 207, Zhoumai 36, Zhoumai 22, Zhengmai 366, Xinong 388, Pingan 518, Saidmai 5, Xinmai 36, Aikang 58 and other wheat varieties widely popularized in the production of Huanghuai wheat region and the germplasm China Spring commonly used in scientific research.
[0035] Wheat backbone germplasm Zhou 8425B is a new type of 1BL / 1RS translocation wheat germplasm created by the distant hybridization of triticale and common wheat varieties through stepwise compound hybridization, radiation mutagenesis and chromosome detection. Firstly, the hexaploid triticale Guangmai 74 was crossed with the common wheat Lianfeng 1, and its F 1 Further crossing with the common 1...
Embodiment 2
[0055] In March 2020, 74 derivative varieties of Zhou 8425B were molecularly detected by the method of the present invention, and their resistance to stripe rust and leaf rust was predicted. The detailed steps of detection are as follows:
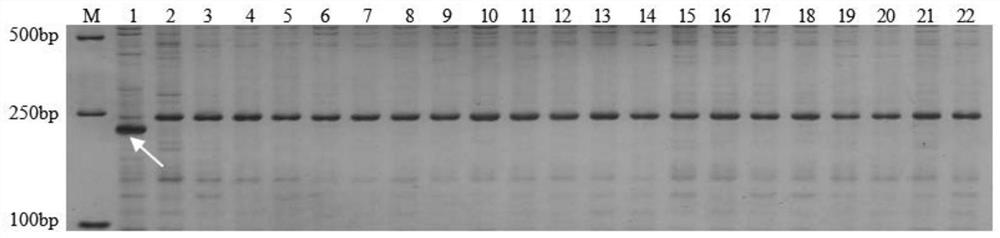
[0056] a. Sample collection: At the jointing stage of wheat, take 184 weekly 8425B derivative varieties with 1.5cm long leaves, put them into sterilized 1.5mL centrifuge tubes, put them in the ice box and bring them back to the laboratory for DNA extraction;
[0057] b. DNA extraction: use the CTAB method (Wang Guanlin, Fang Hongyun. Principles and Technology of Plant Genetic Engineering. Science Press, 1998: 370-372) to extract the DNA of the sample in step a,
[0058] ① After grinding the leaves with liquid nitrogen, quickly add 600 μL CTAB extract to the centrifuge tube, mix well, place in a water bath at 65°C for 1 hour, and shake well every 15 minutes;
[0059] ②Take out the sample and cool it on ice, add 600 μL chloroform / isoamyl alco...
Embodiment 3
[0077] Example 3: The present invention can predict the incidence rate in advance, do hybridization one year in advance, and improve breeding efficiency. The detection material is BC 1 Population of 45 individuals, BC 1 The population was obtained by crossing the high-yielding variety Zhoumai 36 (female parent) with the main parent germplasm Zhou 8425B (male parent) and backcrossing once. 45 BC 1 Individual plants are defined as ZM36 / Z84-1~ZM36 / Z84-45.
[0078] The basic detection steps are basically the same as in Example 2, except that:
[0079] In step b: ⑥Dry the DNA precipitate, add 100 μl of ultrapure water, let stand at room temperature for 60 minutes, and store at -20°C for later use.
[0080] 45 BC planted in March 2021 1 Molecular testing was carried out on individual plants, and the test results are shown in Table 2. In April 2021, 14 individual plants carrying the Z84-1B-1 marker were screened for backcrossing with the high-yielding reincarnation parent Zhoumai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com