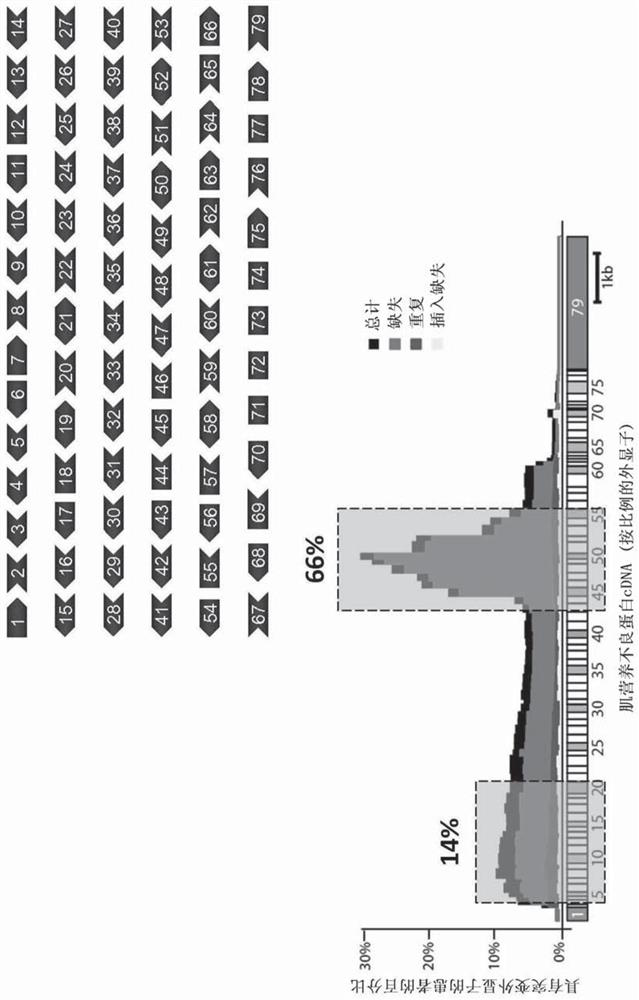
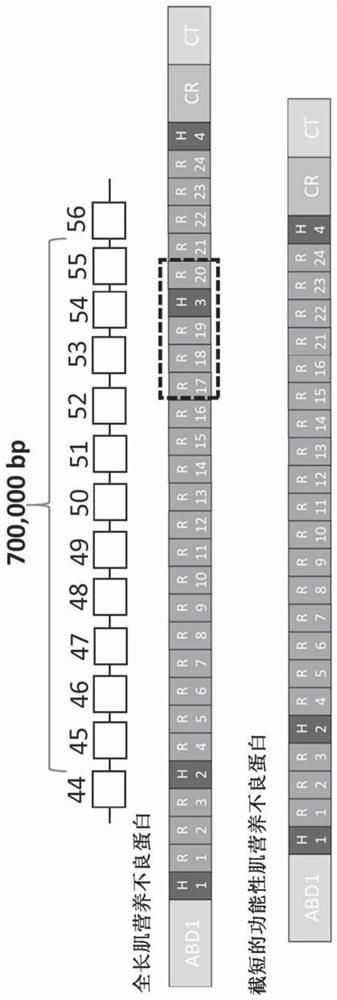
Aav vector-mediated deletion of large mutational hotspot for treatment of duchenne muscular dystrophy
A muscular dystrophy, carrier technology, applied in the fields of genome engineering and genome modification, genome engineering and genome modification of genes in muscles, and can solve the problems of large dystrophin gene sequence, non-delivery, safety issues, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
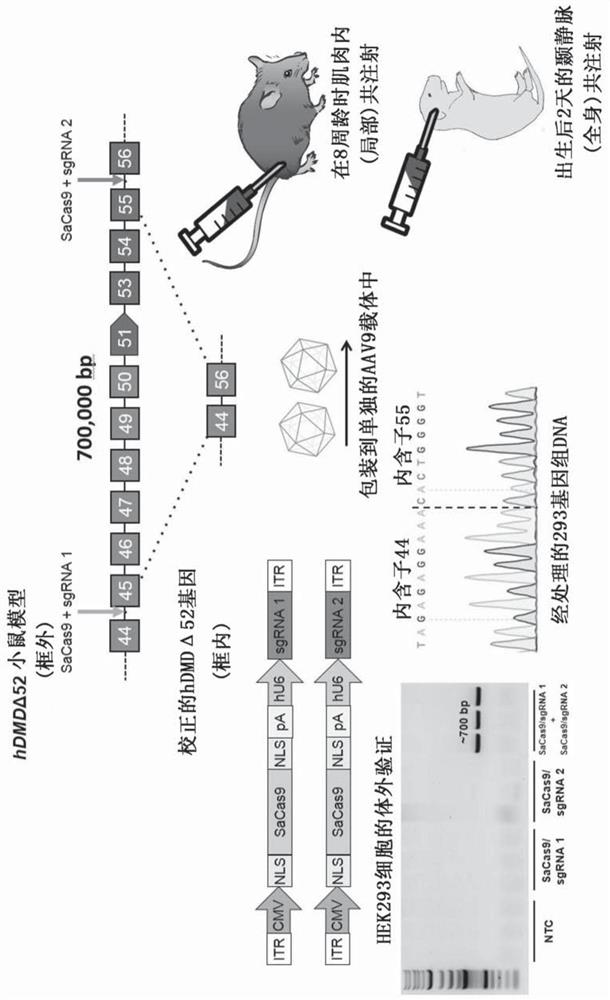
[0192] dual carrier system
[0193] Conventional CRISPR / Cas9 systems for the treatment of DMD typically include more than one vector ( Figure 6 , Figure 7 ). For example, one vector can encode the Cas9 protein, and a second vector can encode two gRNAs. As another example, one vector can encode a Cas9 protein and a first gRNA, and a second vector can encode a Cas9 protein and a second gRNA.
[0194] A schematic diagram of the experiment using multiple vectors to excise exons 45-55 of mouse dystrophin is shown in image 3 , the results are shown in Figure 4 , Figure 5 and Figure 10 middle. Neonatal mice were treated with the dual vector system via systemic / temporal vein injection. Eight weeks after treatment, tissues were harvested. Such as Figure 4 As shown, PCR and sequencing confirmed the deletion of mutation hotspot exons 45–55. Other results are shown in Figure 10 In , AAV-CRISPR targets a control locus ( Figure 10 , upper panel) or targeting exons 45-5...
Embodiment 2
[0196] Validation of the Dual Vector System Therapy Approach
[0197] The CRISPR-based method of restoring a functional dystrophin gene using the dual vector of Example 1 was additionally validated using immortalized myoblasts isolated from DMD patients. Immortalized myoblasts contain a deletion of exons 48-50, creating an out-of-frame mutation ( Figure 9A ). Patient myoblasts were transfected with the same AAV plasmid as used in the HEK293 in vitro experiments in Example 1.
[0198] Deletion PCR of genomic DNA and cDNA revealed efficient deletion of exons 45-55, which was confirmed by Sanger sequencing ( Figure 9B ). Western blot of cell lysates showed that untreated myoblasts did not produce Dystrophin, whereas transfected myoblasts expressed smaller Dystrophin compared to the positive control, consistent with the loss of hotspots ( Figure 9C ). These results also provide in vitro validation that the dual vector construct can be used to edit human mutations and resto...
Embodiment 3
[0200] Components of an all-in-one carrier
[0201] developed a single-vector CRISPR / Cas9 system for the treatment of DMD ( Figure 6 , Figure 7 ). Advantages of single-vector systems may include: having all necessary editing components on a single vector, ability to increase effective dose, simplification of production of other vectors (single therapeutic agent), muscle-specific promoters (e.g., CK8, Spc512, MHCK7) The use / incorporation, and the ability to target combinations of exons and large-scale deletions (e.g., by altering guide sequences). A schematic diagram of the developed all-in-one vector is shown in Figure 8 middle. The sequences contained in some or all of the all-in-one vectors described herein are shown in Table 1. Figure 12 , Figure 13 and Figure 14 The results of testing these constructs in mdx mice are shown. All-in-one vectors are further detailed in Examples 4-7.
[0202]
[0203]
[0204]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com