Screening method of SARS-CoV2 potential mutation sites and application thereof
A mutation site and screening method technology, applied in the fields of bioinformatics and biomedicine, can solve problems such as limited software functions, unsuitability for mutation analysis of large-scale data sets, and inability to realize virus mutation monitoring, etc., to achieve improved packaging efficiency and purity High and stable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Screening method for potential mutation sites of SARS-CoV2
[0041] (1) For a specific new type of coronavirus SARS-CoV2, the whole genome sequencing data of the new crown is uploaded to the GISAID database (https: / / www.gisaid.org / ), and can be downloaded from the GISAID database for free. Use BioAide software to quickly annotate and compare the downloaded sequences, and extract the sequences of all coding genes from the whole genome sequence.
[0042] Sequence comparison: first drag the unaligned virus genome sequence directly to the input box, or load the file path through the file button;
[0043] Then select the sequence type as nucleotide (Nucleotide), the output format as fasta format, and select the alignment strategy as automatic mode (auto);
[0044] Finally, according to the configuration of the computer, select the number of threads to use, and then click the Run (Start) button to complete the comparison of the whole genome of the virus sequence se...
Embodiment 2
[0088] Example 2 Single point mutation example: S-S477N
[0089] Test materials: pcDNA3.1-SARS2-S-HA (diluted to 10 ng / µL), KOD high-fidelity enzyme KOD –Plus-Neo (Code: KOD-401), Dpni enzyme (NEB R0176S)
[0090] Primer design:
[0091] Table 4
[0092]
[0093] Design principles of point mutation primers:
[0094] 1. The length of the mutant primer is 35-40bp. Centering on the amino acid to be mutated, the forward and reverse primers have an overlapping region of 15-20bp.
[0095] 2. The GC content of the mutant primers is between 40-60%, especially at the 3' end.
[0096] The PCR amplification system is shown in Table 5:
[0097] table 5
[0098]
[0099] Mix and centrifuge gently.
[0100] The PCR amplification program is shown in Table 6:
[0101] Table 6
[0102]
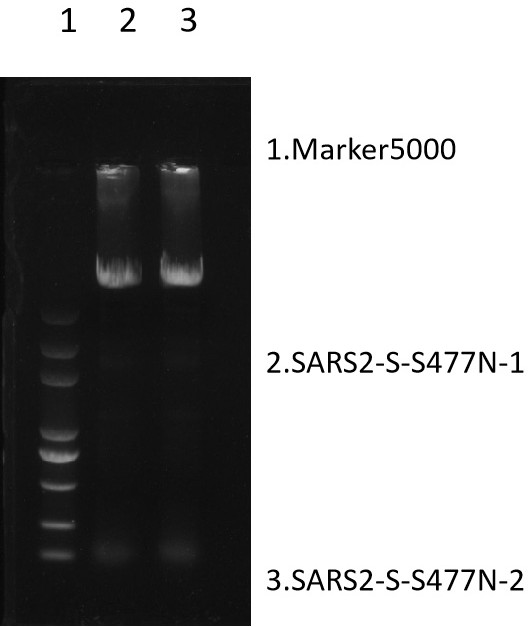
[0103] PCR product identification:
[0104] Prepare 1% agarose gel and spot 5µL PCR product for observation. DNA marker 5000, the target band size is about 9kb. Result: the stripe size con...
Embodiment 3
[0112] Example 3 Example of multiple point mutation: SARS2-S-D614G+D1084Y
[0113] First make a single point mutant SARS2-S-D614G; then add the mutation site D1084Y on this basis.
[0114] Test materials: pcDNA3.1-SARS2-S-HA (diluted to 10 ng / µL), KOD high-fidelity enzyme KOD –Plus-Neo (Code: KOD-401), Dpni enzyme (NEB R0176S)
[0115] Primer design:
[0116] Table 8
[0117]
[0118] The PCR amplification system is shown in Table 9:
[0119] Table 9
[0120]
[0121] Mix and centrifuge gently.
[0122] The PCR amplification program is shown in Table 10:
[0123] Table 10
[0124]
[0125] PCR product identification:
[0126] Prepare 0.8% agarose gel and spot 5µL PCR product for observation. DNA marker 5000, the target band size is about 9kb. The result is as image 3 .
[0127] Dpni enzyme digestion: if there is a band in the identification result SARS2-S-D614G-1, add the remaining 15 µL of the PCR product to 0.4 µL of Dpni enzyme, mix well and place it at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com