CrRNA and CRISPR-Cas12a system for carbapenemase detection and application
A carbapenemase and protein technology, applied in the field of CRISPR-Cas12a system and CrRNA, can solve the problems of complicated operation, long time required, difficult to carry out in ordinary laboratories, etc., and achieves low cost and shortened detection time. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] A CrRNA used for carbapenemase detection in this embodiment.
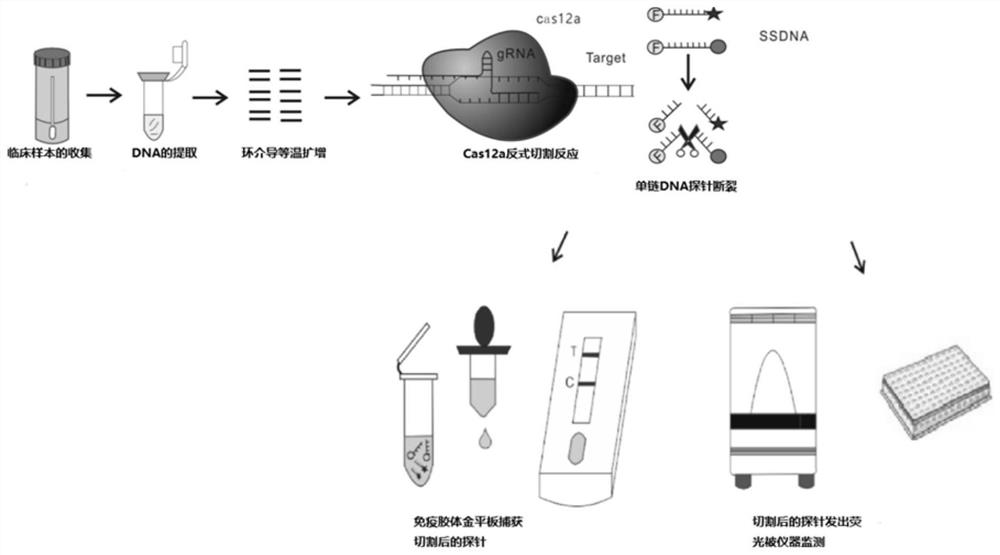
[0040] The core of the CRISPR-Cas12a detection method lies in CrRNA, so the selection of CrRNA is directly related to the effectiveness of the final detection method.
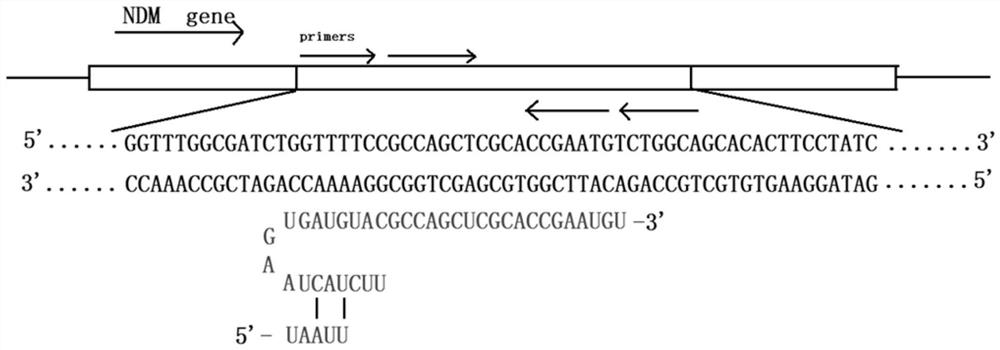
[0041] The base sequence of CrRNA in this example is shown in SEQ ID NO.1, and the design idea is shown in figure 1(The design of crRNA is obtained according to the 20 bases behind the PAM TTTN site recognized by Cas12a). The CrRNA is used to guide the Cas12a protein to recognize and bind to the sequence amplified by LAMP to cut the target sequence. At the same time, the Cas12a protein trans-cuts any single-stranded DNA in the reaction system.
[0042] Specifically, any single-stranded DNA in the reaction system is mainly the LAMP amplification primer in the CRISPR-Cas12a system, including primer F3, primer B3, primer BIP, primer FIP, primer LB, and primer LF, and their sequences are as follows: Shown in SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO....
Embodiment 2
[0044] A CRISPR-Cas12a system for detecting carbapenemase in this embodiment includes Cas12a protein, crRNA, fluorescein isothiocyanate-quencher double-labeled probe, and LAMP amplification product. Wherein, the amplification primers of the LAMP amplification product include: primer F3, primer B3, primer BIP, primer FIP, primer LB, primer LF, the sequences are respectively as SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO.4, Shown in SEQ ID NO.5, SEQ ID NO.6, SEQ ID NO.7.
[0045] At the same time, the CRISPR-Cas12a system also includes the reagents required for the LAMP amplification system and the Cas12a cleavage reaction system.
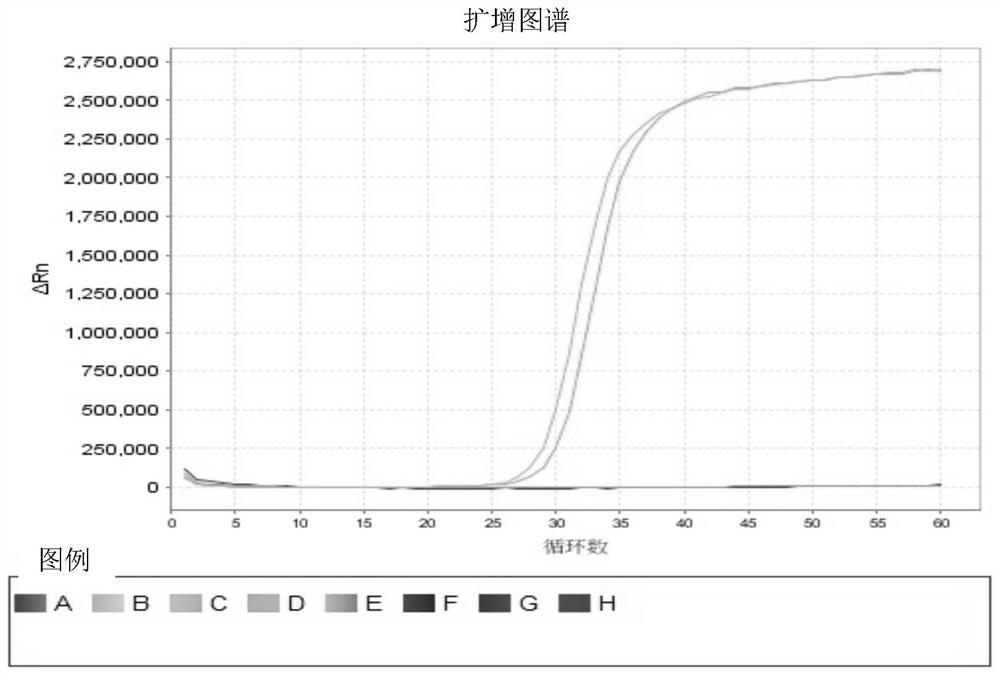
[0046] Specifically, the LAMP amplification system is as follows: primer F3 (0.2 μM), primer B3 (0.2 μM), primer BIP (1.6 μM), primer FIP (1.6 μM), primer LB (0.4 μM), primer LF (0.4 μM) , 1.4mM dNTP, 6mM MgSO 4 , 1× isothermal amplification buffer, 320U / mL Bst 2.0 warm-start DNA polymerase, fluorescein isothiocyanate, 2 μL target DNA template, and add ddH...
Embodiment 3
[0049] A method for the rapid detection of carbapenemase based on CRISPR-Cas12a of this embodiment, using the CRISPR-Cas12a system of Example 2 to detect the sample to be tested, the method is as follows:
[0050] 1. Extraction of DNA from samples to be tested:
[0051] The sample DNA was crudely extracted by boiling method and used as the DNA template for the subsequent LAMP reaction.
[0052] 2. Primer design and LAMP amplification:
[0053] (1) Use primer explorerV5 to design the nucleotide sequence LAMP primer and CrRNA of the target gene fragment NDM and send it to the company for synthesis. Wherein, the target gene fragment NDM is the nucleotide sequence starting from the 563 position of the carbapenemase NDM gene corresponding to the 5' end, and the outer primers F3 and B3 sequences (5'-3') of the nucleotide sequence As shown in SEQ ID NO.2 and SEQ ID NO.3, the sequences (5'-3') of the internal primers FIP and BIP are shown in SEQ ID NO.2 and SEQ ID NO.3.
[0054] (2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com