Primer group for detecting mycoplasma hyopneumoniae, application of primer group and real-time fluorescent quantitative PCR detection method
A real-time fluorescence quantitative technology for Mycoplasma hyopneumoniae, applied in the biological field, can solve problems such as fluctuation of results, identification, counting errors, time-consuming and labor-intensive, etc., and achieve rapid detection, accurate quantification, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
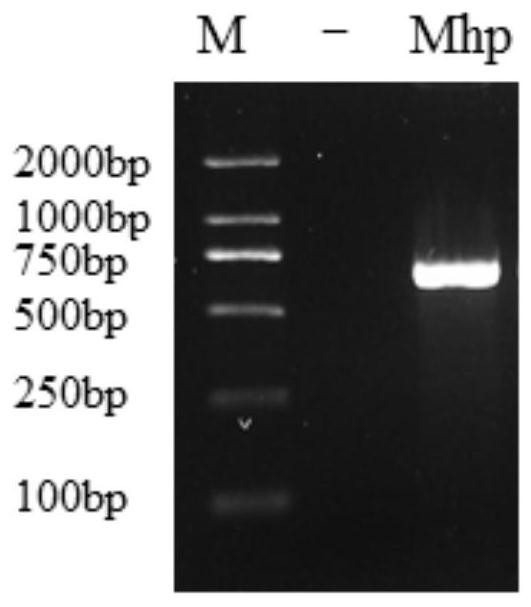
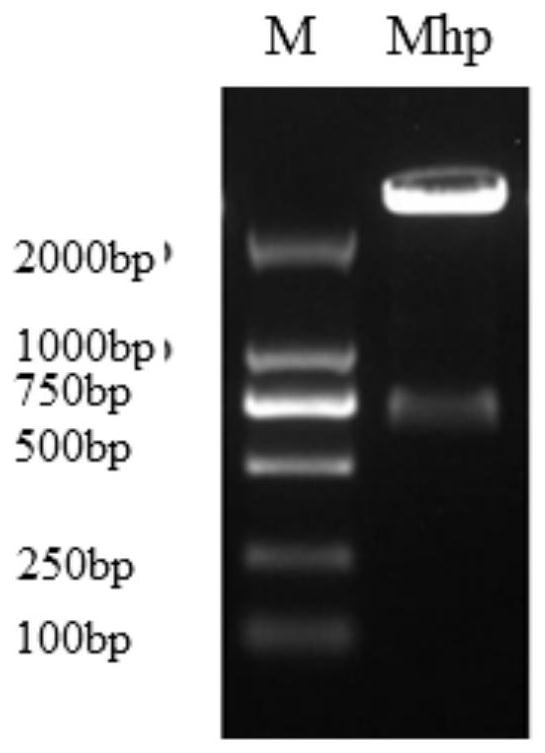
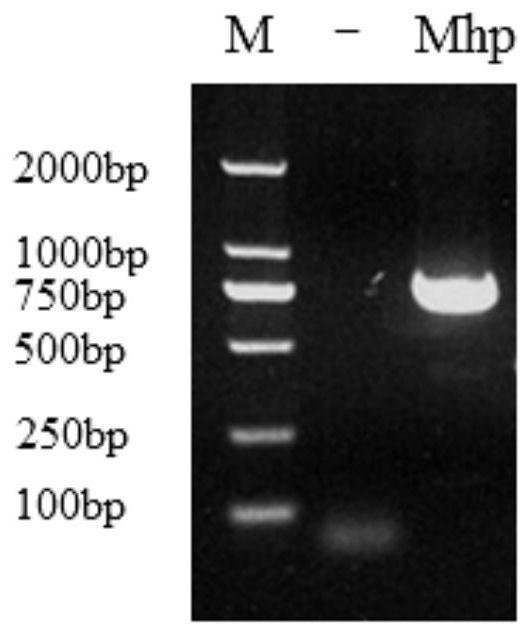
Image
Examples
Embodiment 1
[0059] 1 Primer and probe design and synthesis
[0060] Aiming at the highly conserved region of the 16S rRNA gene genus, which is specific and the main target gene for bacterial classification and identification, as the amplification region, the first primer pair realtime F / R for Real Time PCR was designed, and the second primer pair clone for constructing a standard plasmid was designed F / R, the sequences of the primers and probes are:
[0061] realtime F: 5'-AACTGGTCATATATTGACACTAAGGG-3' (SEQ ID NO.1);
[0062] realtime R: 5'-TGTGTTAGTGACTTTTGCCACCAACT-3' (SEQ ID NO. 2).
[0063] Probe: 5'-CAGGATTAGATACCCTGGTAGTCCAC-3' (SEQ ID NO.3);
[0064] clone F: 5'-GAGCCTTCAAGCTTCACCAAGA-3' (SEQ ID NO.4);
[0065] clone R: 5'-GCGGATCATTTAACGCGTTAGC-3' (SEQ ID NO. 5).
[0066] The product of the primer pair realtime F / R is located within the sequence amplified by the primer pair clone F / R. The 5' end of the Real Time PCR probe is bound with a fluorescent reporter group (FAM), and ...
Embodiment 2
[0093] Comparison of the results of detection of Mycoplasma hyopneumoniae by CCU method and fluorescent quantitative PCR method
[0094] 1 Culture of Mycoplasma hyopneumoniae
[0095] Take out the production seeds from the strain bank, dissolve them with the improved Friis liquid medium, and subculture in the improved Friis liquid medium at a ratio of 1:10, put them into a constant temperature incubator at a temperature of 36-38°C and cultivate them for 48-72 hours, and wait for The color of the bacterial liquid changes, and when the pH value drops to 6.70-6.80, the bacterial liquid is collected and used as a first-class seed liquid for pure inspection. It should be pure. Propagate and identify the secondary and tertiary seeds according to the above method; add the tertiary seed liquid into the improved Friis liquid medium at a ratio of 1:10, seal the mouth of the fermenter, feed sterile air, and the pressure in the tank reaches 0.08MPa , close all the inlets and outlets on t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com