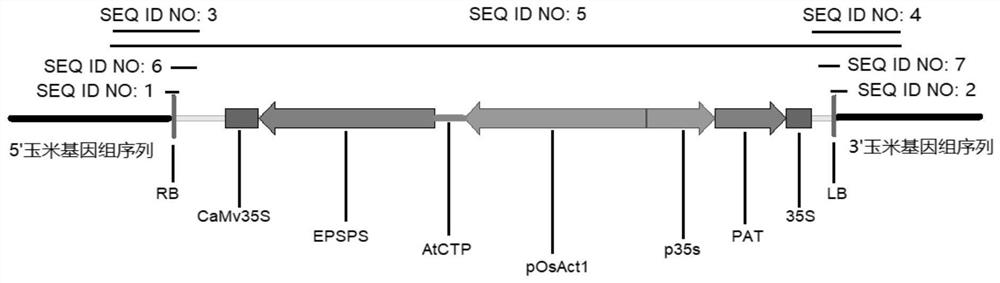
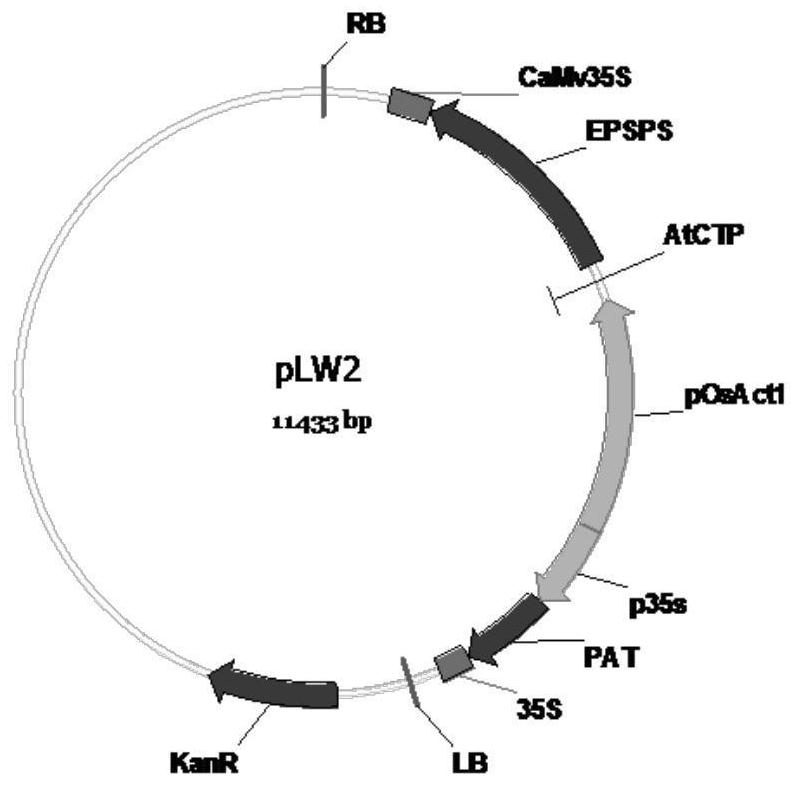
Transgenic maize event LW2-2 and detection method thereof
A technology for transgenic maize and maize events, which is applied in the field of molecular biology to achieve the effects of excellent performance, enhanced breeding efficiency and high tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0164] Embodiment 1 cloning and transformation
[0165] 1.1. Vector cloning
[0166] Use standard gene cloning techniques to construct recombinant expression vector pLW2 (such as figure 2 shown). The vector pLW2 comprises two transgenic expression cassettes in series, the first expression cassette is operably linked to the coding sequence (spAtCTP2) of the Arabidopsis EPSPS chloroplast transit peptide by the rice actin 1 promoter (pOsAct1), which can operably linked to the glyphosate-tolerant 5-enol-pyruvylshikimate-3-phosphate synthase (cEPSPS) of the Agrobacterium sp. CP4 strain, operably linked to the transcription terminator of cauliflower mosaic virus 35s ( CaMv35S); the second expression cassette consists of a tandem repeat of the cauliflower mosaic virus 35S promoter (p35S) containing an enhancer region operably linked to the glufosinate-tolerant phosphinothricin of Streptomyces N-acetyltransferase (cPAT) and is operably linked to cauliflower mosaic virus 35S termin...
Embodiment 2
[0174] Example 2 Detection of transgenic maize event LW2-2 with TaqMan
[0175] About 100mg of the leaves of the transgenic maize event LW2-2 were taken as a sample, and the genomic DNA was extracted with Qiagen's DNeasyPlantMaxiKit, and the copy numbers of the EPSPS gene and the PAT gene were detected by fluorescent quantitative PCR with Taqman probes. At the same time, wild-type maize plants were used as a control, and detection and analysis were carried out according to the above method. The experiment was repeated 3 times, and the average value was taken.
[0176] The specific method is as follows:
[0177] Step 11, take 100 mg of leaves of transgenic corn event LW2-2, grind into a homogenate with liquid nitrogen in a mortar, and take 3 replicates for each sample;
[0178] Step 12, using the DNeasy Plant Mini Kit of Qiagen to extract the genomic DNA of the above sample, the specific method refers to its product manual;
[0179] Step 13, measure the genomic DNA concentra...
Embodiment 3
[0196] Example 3 Detection of transgenic corn event LW2-2
[0197] 3.1. Genomic DNA extraction
[0198] The DNA was extracted according to the conventional CTAB (cetyltrimethylammonium bromide) method: after taking 2 grams of tender leaves of the transgenic corn event LW2-2 and grinding them into powder in liquid nitrogen, add 0.5mL at a temperature of 65 ℃ preheated DNA extraction CTABBuffer (20g / L CTAB, 1.4M NaCl, 100mM Tris-HCl, 20mM EDTA (ethylenediaminetetraacetic acid), adjust the pH to 8.0 with NaOH), mix thoroughly, and pump at a temperature of 65℃ Extract for 90 minutes; add 0.5 times the volume of phenol and 0.5 times the volume of chloroform, mix upside down; centrifuge at 12,000 rpm (revolutions per minute) for 10 minutes; absorb the supernatant, add 2 times the volume of absolute ethanol, and gently shake the centrifuge tube. Let stand at 4°C for 30 minutes; centrifuge at 12,000 rpm for 10 minutes; collect DNA to the bottom of the tube; discard the supernatant an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com