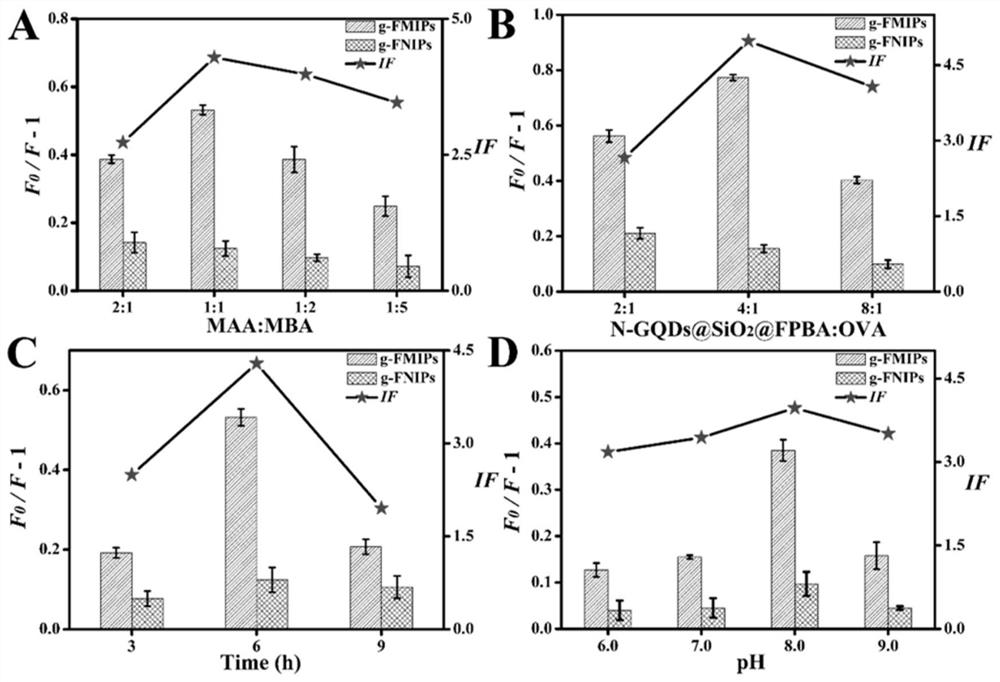
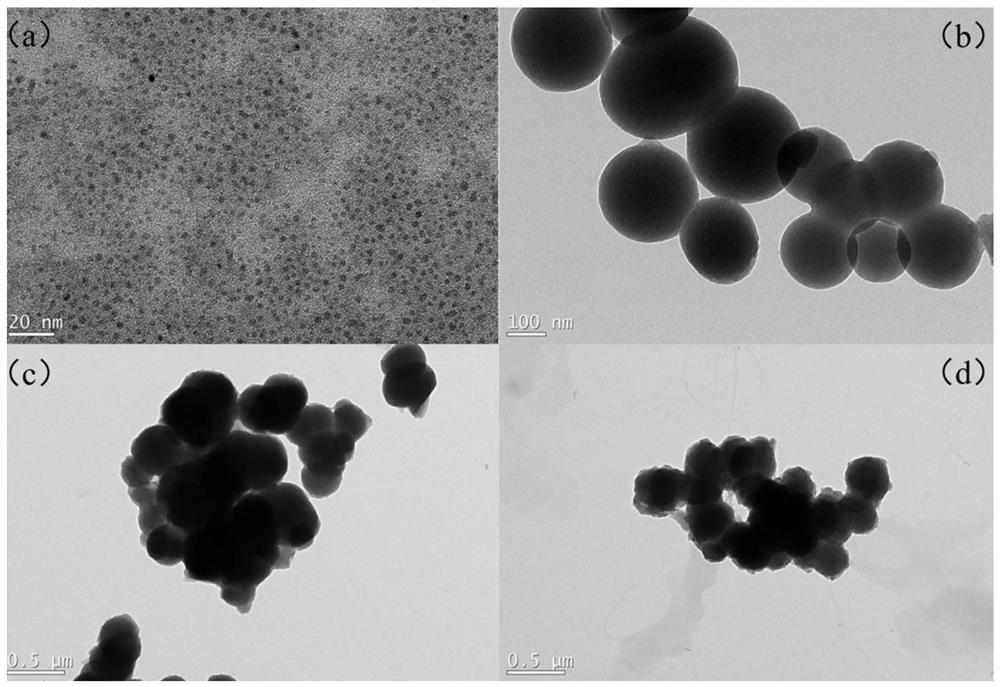
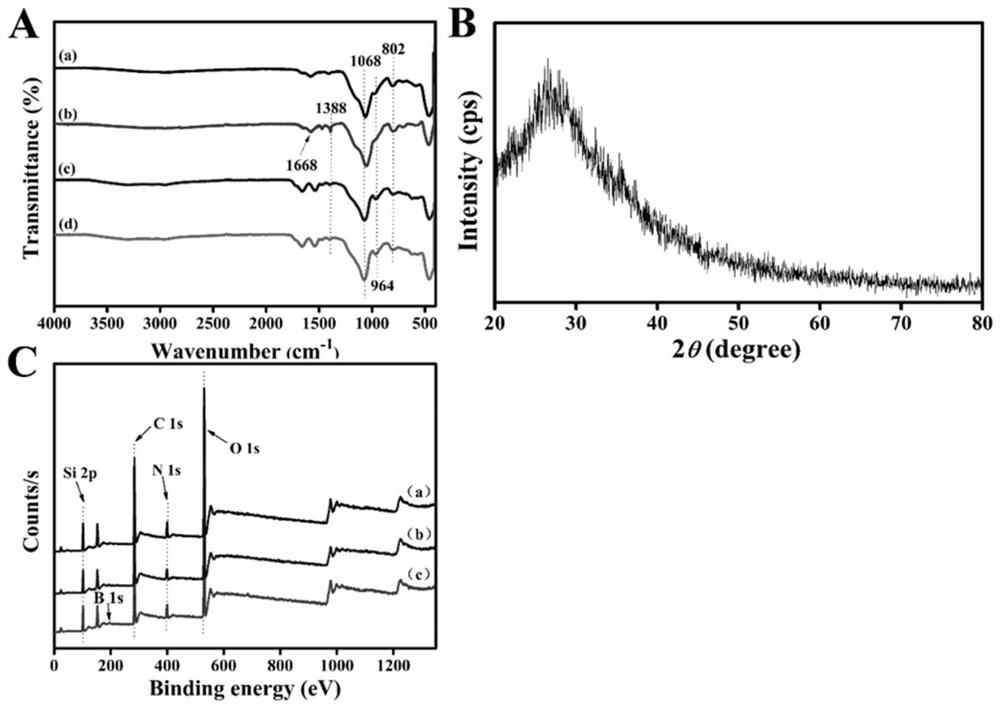
Fluorescent molecularly imprinted polymer based on glycopeptide, preparation method and application in glycoprotein screening and detection
A fluorescent molecular imprinting and polymer technology, applied in the field of biosensing and bioanalysis, can solve the problems of complex structure, variability, large molecules, etc., and achieve the effect of improving specificity, binding force and good photostability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0042] Preparation of water-soluble N-GQDs. Dissolve CA (0.22-0.23 g) and EDA (0.18-0.22 mL) in ultrapure water (4.5-5.5 mL), and stir for 10-30 minutes until clear. The solution was transferred to a 20mL polytetrafluoroethylene reactor, and reacted at 160-200°C for 6-10h. Excess absolute ethanol was added to the obtained product to obtain solid N-GQDs.
[0043] In order to prepare N-GQDs@SiO 2 For nanoparticles, add N-GQDs (10-12mL), TEOS (20-60μL) and APTES (20-60μL) into absolute ethanol (30-50mL) under magnetic stirring, and react at room temperature for 1-3h. After washing three times with absolute ethanol, it was dried with a freeze dryer.
[0044] Subsequently, for the N-GQDs@SiO 2 Amino groups and double bonds are modified on the surface of nanoparticles, and N-GQDs@SiO 2 (790-810mg), anhydrous toluene (10-11mL), APTES (5-6mL) and MPS (5-6mL) were put into a round bottom flask, sonicated for 30min to obtain a suspension. the suspension in N 2 Reflux at 90-120°C ...
Embodiment 1
[0052] 1. Materials and methods
[0053] 1. Experimental materials
[0054] OVA, bovine serum albumin (BSA), bovine hemoglobin (BHb) and pronase E (Pronase E) (Beijing Suolaibao Technology Co., Ltd.); horseradish peroxidase (HRP) and trypsin (Tryspin) ( Shanghai Sangon Bioengineering); α-methacrylic acid (MAA), 3-aminopropyltriethoxysilane (APTES), 3-methacryloyloxypropyltrimethoxysilane (MPS), tetraethyl Oxysilane (TEOS), tetramethylethylenediamine (TEMED) and N, N'-methylenebisacrylamide (MBA) (Shanghai McLean Biochemical Technology Co., Ltd.); 4-formylphenylboronic acid (4- FPBA) (Shanghai Aladdin Biochemical Technology Co., Ltd.); ethylenediamine (EDA) and sodium borohydride (NaBH 4 ) (Shaanxi Sinopharm Holding Chemical Reagent Co., Ltd.); ammonium bicarbonate (NH 4 HCO 3 ), ammonium persulfate (APS), sodium chloride (NaCl) and sodium dodecyl sulfate (SDS) (Guangdong Guanghua Technology Co., Ltd.).
[0055] 2. Preparation of g-FMIPs
[0056] 2.1N-GQDs@SiO 2 Synthesi...
Embodiment 2
[0104] 2.1 N-GQDs@SiO 2 Synthesis of @FPBA nanoparticles
[0105] Preparation of water-soluble N-GQDs. CA (0.22 g) and EDA (0.18 mL) were dissolved in ultrapure water (4.5 mL), stirred for 10 minutes until clear. The solution was transferred to a 20mL polytetrafluoroethylene reactor, and reacted at 200°C for 6h. Excess absolute ethanol was added to the obtained product to obtain solid N-GQDs.
[0106]In order to prepare N-GQDs@SiO 2 For nanoparticles, N-GQDs (11 mL), TEOS (20 μL) and APTES (20 μL) were added into absolute ethanol (30 mL) under magnetic stirring, and reacted at room temperature for 3 h. After washing three times with absolute ethanol, it was dried with a freeze dryer.
[0107] Subsequently, for the N-GQDs@SiO 2 Amino groups and double bonds are modified on the surface of nanoparticles, and N-GQDs@SiO 2 (790mg), anhydrous toluene (10mL), APTES (5mL) and MPS (5mL) were put into a round bottom flask and ultrasonicated for 30min. the suspension in N 2 Refl...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com