Optimized expression of nitrilase promoter and application thereof
A nitrilase and promoter technology, applied in hydrolytic enzymes, enzymes, biochemical equipment and methods, etc., can solve the problems of inability to alleviate inclusion body production, high expression yield, inclusion body precipitation, etc., and achieve large-scale industrial application prospects , mild conditions, good selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1: construction of nitrilase genetically engineered bacteria
[0026] The nitrilase of the present invention is obtained by the method of gene cloning, adopts bacterial genome extraction kit (Takara) to extract A.facilis ATCC11228 genome as template, according to NCBI database A.facilis nitrilase homology analysis, design primer pair nit The gene is amplified by PCR, and the primer sequences are shown in SEQ ID NO:3 and SEQ ID NO:4. The PCR system (100 μL) is: template (10ng / μL) 0.5 μL, upstream primer (50 μM) 0.5 μL, downstream primer (50 μM) 0.5 μL, dNTPMixture (2.5mM) 8 μL, 5×PrimeSTAR Buffer 20 μL, ddH 2 O 69.5 μL, PrimeSTAR DNA polymerase 1 μL. PCR program: (1) Denaturation at 98°C for 3 minutes, (2) Denaturation at 98°C for 10 seconds, (3) Annealing at 60°C for 5 seconds, (4) Extension at 72°C for 1.2 minutes, cycle (2)-(4) steps 30 times, (5 ) at 72°C for 5 min.
[0027]Take the PCR product for gel recovery to obtain the target gene, use NcoI and Xh...
Embodiment 2
[0028] Example 2: Construction of recombinant strains containing different promoters
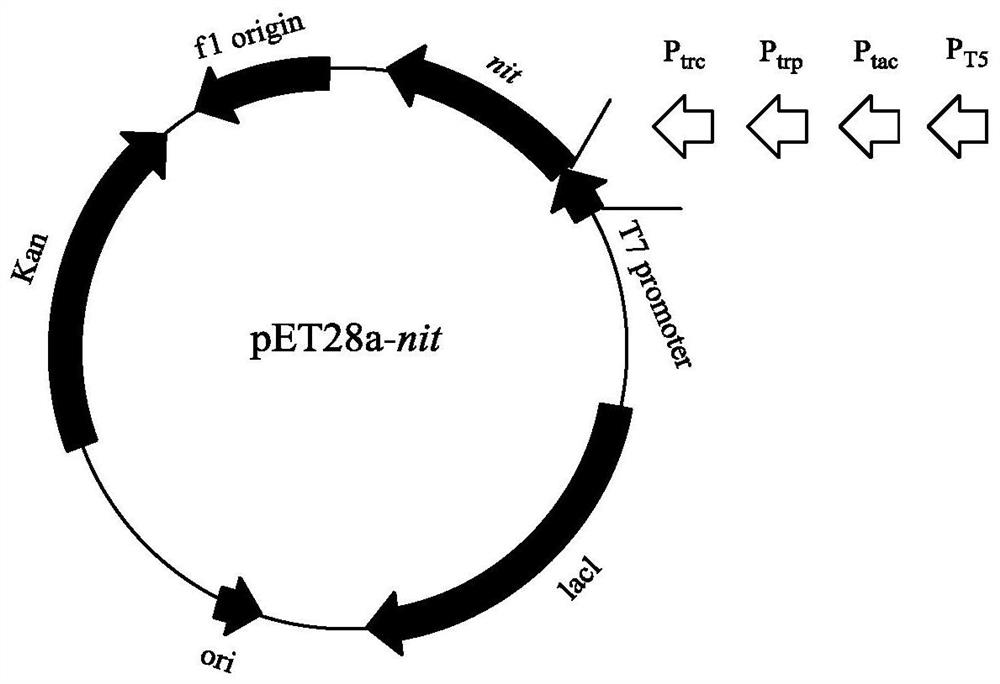
[0029] The recombinant strain E.coli BL21(DE3) / pET28a-nit constructed in Example 1 was cultivated, the plasmid was extracted as a promoter mutation template, and primers were designed to mutate the recombinant plasmid. See SEQ ID NO: 5 to SEQ ID NO: for the primer sequence 12. For the map of recombinant expression plasmid pET28a-T7-nit and the promoter replacement scheme, see figure 1 . The PCR system (100 μL) is: template (10ng / μL) 0.5 μL, upstream primer (50 μM) 0.5 μL, downstream primer (50 μM) 0.5 μL, dNTP Mixture (2.5mM) 8 μL, 5×PrimeSTAR Buffer 20 μL, ddH 2 O 69.5 μL, PrimeSTAR DNA polymerase 1 μL. PCR program: (1) Denaturation at 98°C for 3 minutes, (2) Denaturation at 98°C for 10 seconds, (3) Annealing at 60°C for 5 seconds, (4) Extension at 72°C for 6.5 minutes, cycle (2)-(4) steps 30 times, (5 ) at 72°C for 5 min.
[0030] Take the PCR product for DpnI treatment to digest the ...
Embodiment 3
[0031] Embodiment 3: the preparation of different promoter NIT bacterial cells
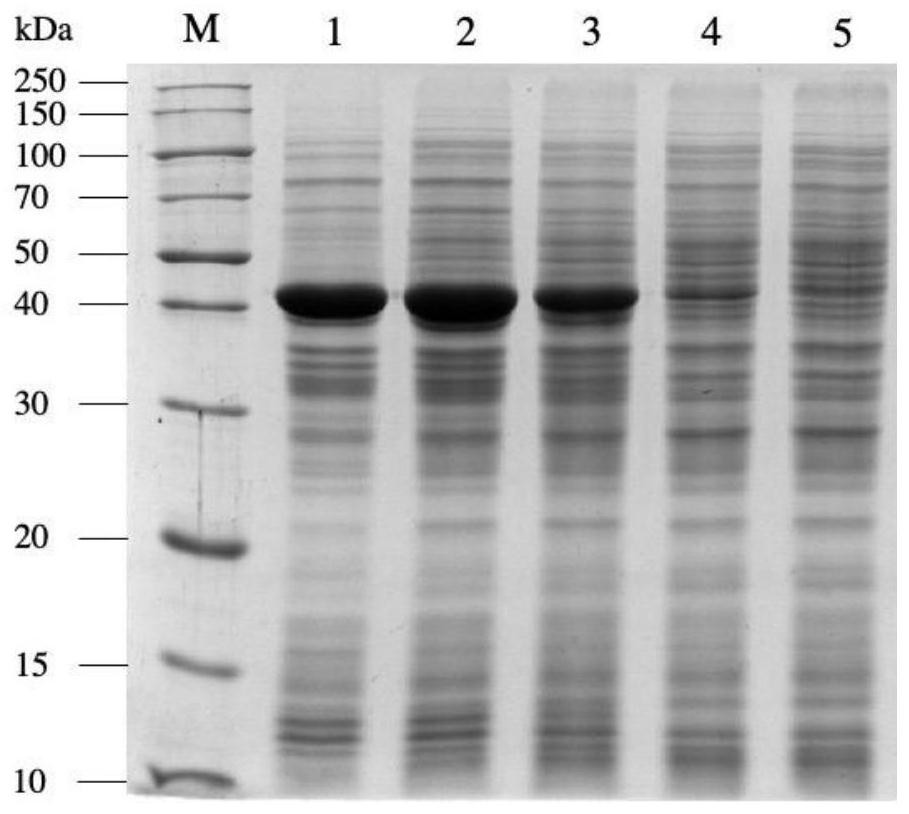
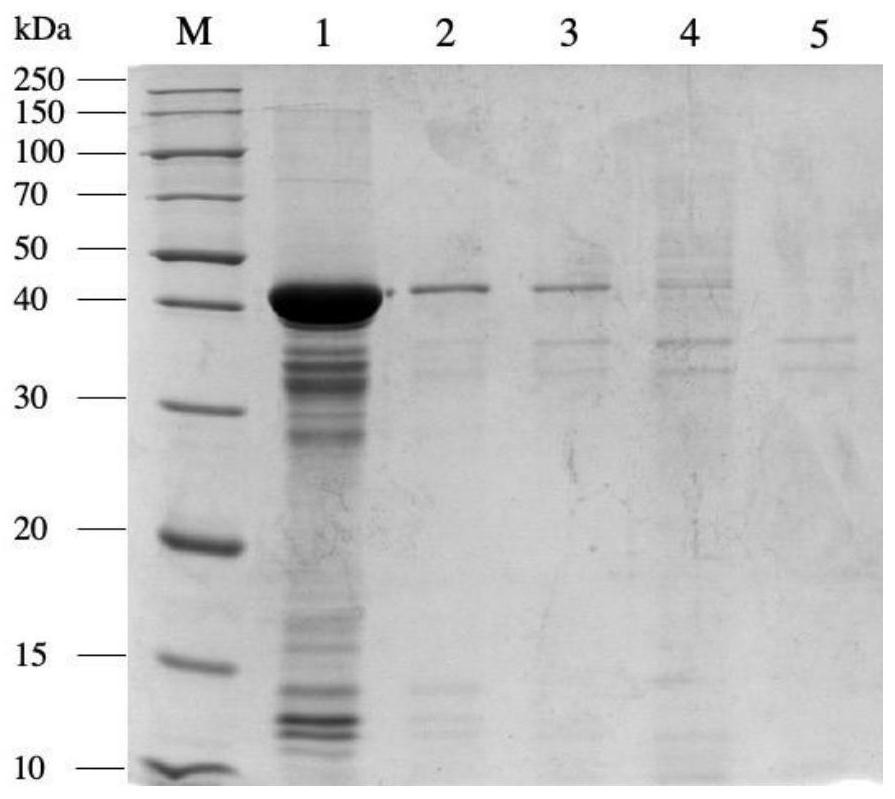
[0032] The genetically engineered bacteria pET28a-nit constructed in Examples 1 and 2 were inoculated into the fermentation medium containing 50 μg / mL kanamycin, and cultivated at 37°C to the cell concentration OD 600 If the value is 0.6-0.8, add a final concentration of 0.1mmol / L IPTG to the fermentation medium, induce culture overnight at 28°C, centrifuge the culture solution at 4°C, 12000rpm for 5min, discard the supernatant, and collect NIT expressed by different promoters wet bacteria. Each weighed 1g of wet bacteria, suspended in 10mL phosphate buffer (pH 7.0), ultrasonically disrupted, and analyzed the supernatant and precipitate of the broken bacteria by SDS-PAGE protein denaturation electrophoresis ( figure 2 and image 3 ), the results showed that the expression of pET28a-T7-nit recombinant protein was the largest, but because the expression rate was too fast, the proportion of inclus...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com