Prokaryotic expression method and application of EqHV NS3 protein
A protein and expression bacteria technology, applied in the biological field, can solve the problems of lack of detection of EqHV antibodies, etc., achieve good stability, promote lymphocyte stimulation, good sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
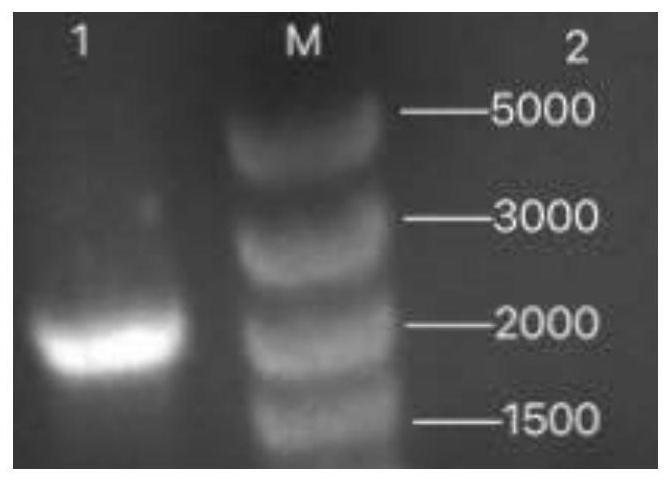
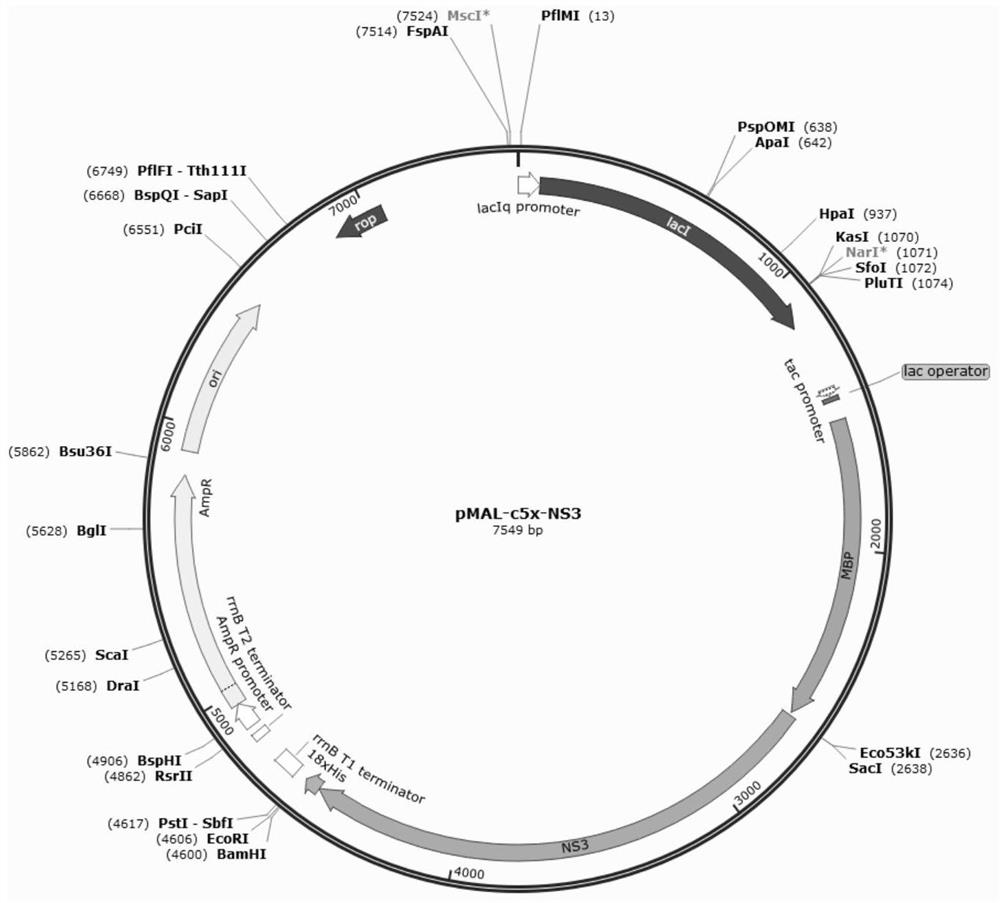
[0069] Example 1: Construction of recombinant expression vector pMAL-c5x-NS3
[0070] (1) Primer design
[0071] According to the NS3 gene (SEQ ID NO: 3) of EqHV strain, design PCR primer, NS3 gene PCR primer name is SacI 1893F and Bam I 1893R, and cDNA is the reverse transcription product of GD18 strain (the Guangdong strain of EqHV), The primer sequence contains Sac I and Bam I restriction sites, protective bases and bases to prevent frameshift (as shown in the underline), and the downstream primers are inserted with 3 6*His and terminators. The primers were provided by Tianyi Synthesized by Huiyuan Biological Co., Ltd., the sequence is as follows:
[0072] Sac I 1893F:
[0073] 5' ATCGAGCTC GATGTCACCTATTACAGCCACTGTCACT 3' (SEQ ID NO: 1)
[0074] Bam I 1893R:
[0075] 5' CGCGGATCC CTATCATTAATGGTGATGGTGATGATGATGGTGATGGTGATGATGATGGTGATGGTGATGATGCGTTTGGGTGTCAATCTCGGCCTCCAT 3' (SEQ ID NO: 2)
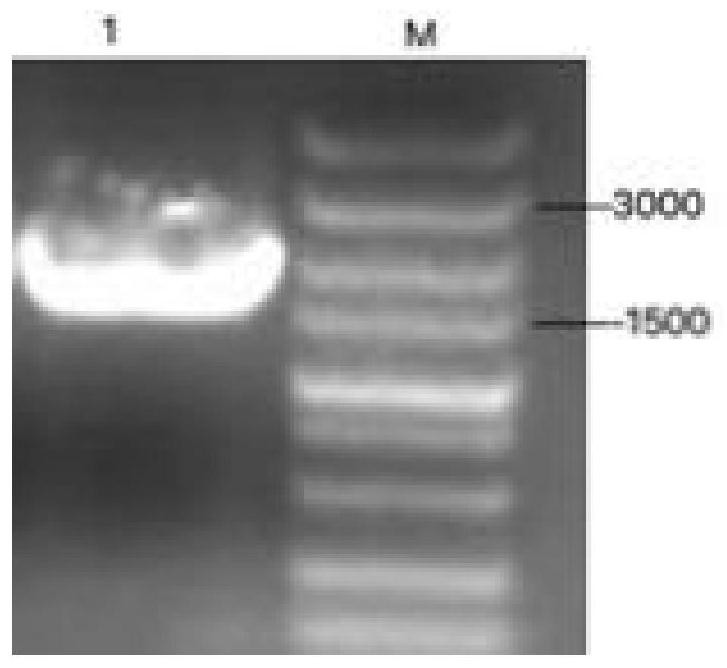
[0076] (2) PCR amplification of NS3 gene
[0077] Use the primers designed ...
Embodiment 2
[0119] Example 2: Expression and purification of recombinant NS3 protein
[0120] 1. Expression of recombinant NS3 protein
[0121] (1) Transform the recombinant expression vector pMAL-c5x-NS3 into the expression strain competent cell Rosetta (DE3), and the specific transformation method is the same as the step of transforming the above-mentioned ligation product into the E. coli DH5α competent cell;
[0122] (2) Use a sterilized 10 μL pipette tip to pick a single colony that grows well on the ampicillin plate, place it in 1 mL of ampicillin-resistant LB broth, place it in a shaker at 37°C, and shake at 180 rpm for 12 to 15 hours;
[0123] (3) Put the shaken bacterial solution into the ampicillin LB medium at a ratio of 1:100, and culture it in a shaker at 200 rpm at 37°C until the bacterial cell OD 600nm When the value is about 0.5, add the inducer IPTG with a final concentration of 1 mM, shake and culture in a shaker at 25°C for 5 hours, and then set aside;
[0124] (4) Ce...
Embodiment 3
[0140] Example 3: Determination of the optimal induction conditions for recombinant NS3 protein expression
[0141] (1) Determination of the optimal induction temperature
[0142] In the step (3) of the expression procedure of the recombinant NS3 protein of embodiment 2, when the bacterium OD 600nm When the value reaches about 0.6, take three bacterial solutions, add the same amount of IPTG to the bacterial solutions, induce expression at 25°C, 30°C and 37°C respectively, and culture at 180rpm for the same time. Centrifuge at 8000rpm for 5min at room temperature, discard the supernatant and retain the cells obtained by centrifugation, resuspend the cells with 0.1 times the original volume of PBS buffer, take the resuspended cells as loading samples for SDS-PAGE electrophoresis, The results of different induction temperatures for recombinant NS3 protein expression are shown in the attached Figure 6 As shown in the figure, M is Marker, 1 is the induction temperature of 25°C, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com