CRISPR system and application thereof to mortierella alpina
A technology of Mortierella alpina and nuclease, applied in the field of bioengineering, can solve problems such as low efficiency and no CRISPR knockout system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
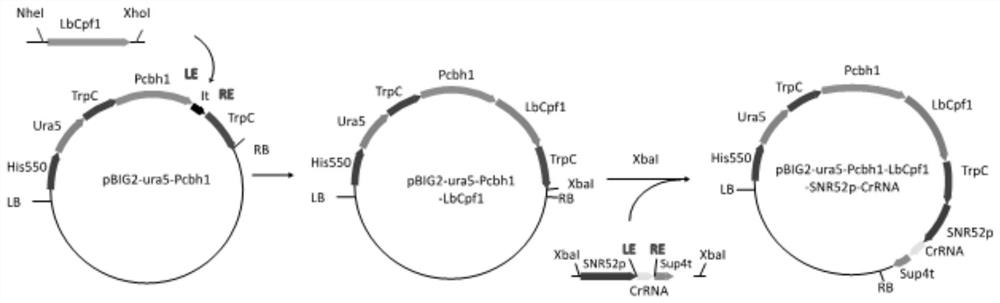
[0046] Example 1 Construction of expression vector pBIG2-Pcbh1-LbCpf1
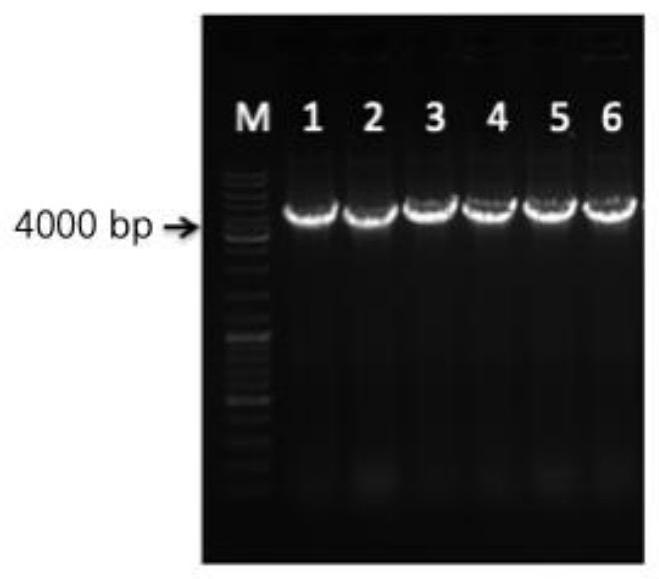
[0047] The selected nuclease is from LbCpf1 of Lachnospiraceae. A SV40 nuclear localization sequence (CCCAAGAAGAAGCGCAAGGTCC) was added at the end of the nuclease sequence. The entire fragment was biosynthesized by GenScript after codon optimization. The nucleotide sequence is shown in SEQ ID NO.1. After the synthesized LbCpf1 nuclease fragment was digested and purified by NheI and XhoI, a high-purity and high-concentration gene fragment was obtained, which was integrated into the binary vector pBIG2 in Mortierella alpina which had also been digested by T4 ligase The plasmid pBIG2-Pcbh1-LbCpf1 was obtained on -ura5-Pcbh1-ITs-TrpCT.
Embodiment 2
[0048] Example 2 Construction of Mortierella alpina Binary Expression Vector pBIG2-ura5-Pcbh1-ITs-TrpCT
[0049] The selected inducible promoter Pcbh1 (shown in SEQ ID NO.2) was derived from Trichoderma reesei, and the sequence was synthesized by GenScript Biotechnology Co., Ltd., and was digested with XbaI and the pBIG2-ura5 plasmid constructed in the laboratory (detailed See the paper "Studies on Transcriptional Regulation and Reducing Power of Mortierella Alpina Fatty Acid Synthesis Process" published in 2015) was constructed as pBIG2-ura5-Pcbh1-ITs-TrpCT. The specific enzyme cleavage reaction and ligation reaction are as follows:
[0050] 1.1 Enzyme digestion reaction: On the basis of the plasmid pBIG2-ura5-ITs, the promoter H550 of the H550-ITs-TrpCT fragment was replaced with Pcbh1 to obtain Pcbh1-IT-TrpCT (for the construction of ITs-TrpCT, see the doctoral thesis "Mortierella alpina Transcriptional Regulation and Reducing Power of Fatty Acid Synthesis Process"). At 3...
Embodiment 3
[0059] Example 3 Construction of expression vector pBIG2-ura5-Pcbh1-LbCpf1-TrpCT-SNR52p-CrRNA
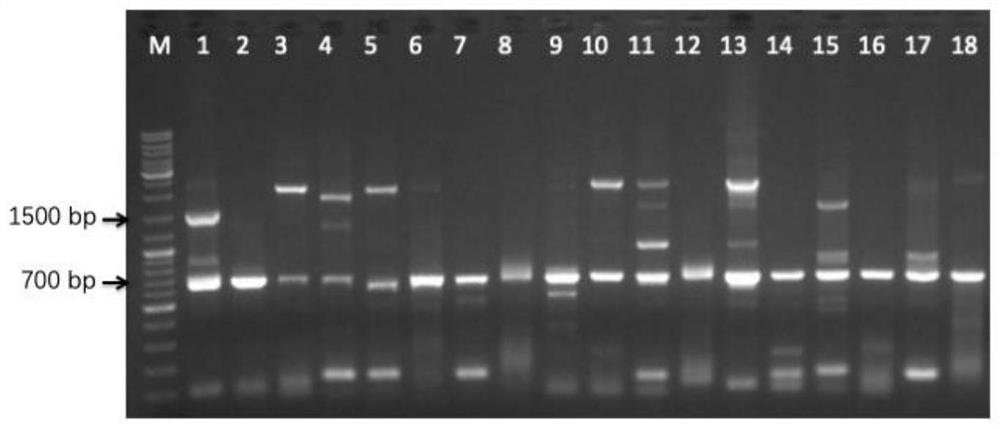
[0060] Design the guide strand CrRNA for the nuclease LbCpf1 targeting gene ura5. In CasOFFinder, the relevant parameters for the guide strand are set as: the operating microorganism is Mortierella alpina ATCC 32222: PAM sequence 5'-3'TTTN of LbCpf1: target The directional gene is ura5 fragment; the length of the guide strand fragment is 23bp. For the core functional conserved region of the ura5 gene, two guide strands that can cover the region to the greatest extent and have a higher score are selected. The specific information is: the off-target score of the guide strand is 62.9, and the CrRNA corresponding to LbCpf1 covers 207bp-231bp of the ura5 fragment. The nucleotide sequence of CrRNA is shown in SEQID NO.4.
[0061] The function of the guide strand of the CRISPR system requires the guidance of RNA polymerase III (polymerase III, pol III) promoters, but there is no relevant...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com