Method and kit for detecting type 2 diabetes mellitus susceptible intestinal bacteria
A technology for type 2 diabetes and intestinal bacteria, applied in the field of microbial detection, to achieve the effect of strong detection specificity, good stability and saving consumables
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Design and screening of embodiment 1 multiplex qPCR primers and probes
[0045] The present invention utilizes multiple fluorescent PCR technology to target the highly conserved region of 16S rRNA of four types of intestinal bacteria susceptible to type 2 diabetes patients, namely, Bacteroides polymorpha, Clostridium tenabilis, Clostridium sphaericus and Lachnospira. Three pairs of specific primers were designed, and two Taqman probes were designed for each strain. The gene accession numbers of the genes used are: CP012937.1, CAJJWU010000001.1, CAJTKH010000001.1, PEDL01000001.1. Through the single-channel primer-probe NTC test and the four-channel primer-probe test, a set of qPCR primers and probes that can be used to simultaneously detect the above four intestinal bacteria and have good detection effects were screened out. The screening process is as follows:
[0046] (1) Single channel primer probe NTC test
[0047] Arrange and combine the designed and synthesized ...
Embodiment 2
[0057] Embodiment 2 Establishment of Multiplex qPCR Detection Method and Detection Kit
[0058] Using the constructed recombinant plasmids of Bacteroides polymorpha, Clostridium tenabilis, Clostridium sphaericus and Lachnospira as templates, the primers and probes screened in Example 1 were used to construct a multiplex qPCR detection method and optimize it.
[0059] Optimization of the reaction system:
[0060] (1) Optimizing the amount of primer probe
[0061] Prepare the primer-probe combination confirmed by screening into a PCR reaction system, adjust the amount of primer probe, prepare multiple systems, and dilute the synthesized plasmid 10 times into 4 gradient concentrations as positive templates for amplification detection , select the appropriate amount of primer probe.
[0062] Table 2 Optimization of the amount of primers and probes for multiplex qPCR
[0063] Material name System 1 System 2 System 3 System 4 System 5 5×DNA PCR Buffer 5...
Embodiment 3
[0083] Sensitivity detection of embodiment 3 multiplex qPCR primers and probes
[0084] Mix the recombinant plasmids described in Example 2 that have been rated as the initial sample, and dilute to a concentration of 10 6 copies / mL, serially diluted to 10 5 、10 4 、10 3 And 500copies / mL as the sample to be tested, test the sensitivity of multiple qPCR primers and probes, the reaction system and reaction conditions are the same as in Example 2.
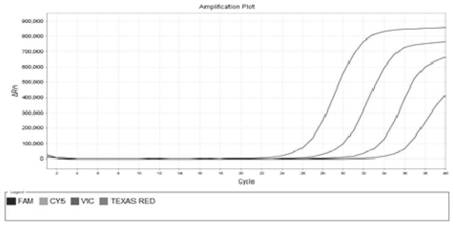
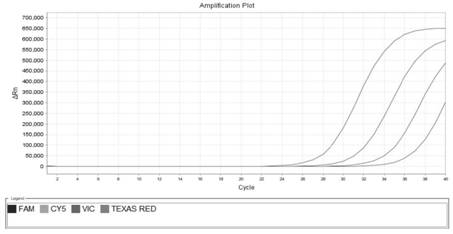
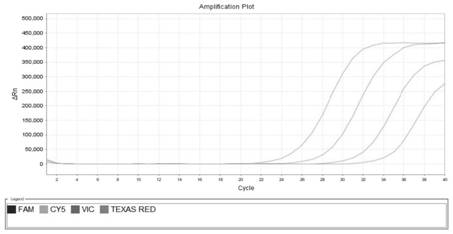
[0085]In order to avoid the influence of the overlapping curves on viewing, the present invention observes the results separately through the fluorescent channels corresponding to the detection probes. The sensitivity test results of qPCR primers and probes are as follows: Figure 1~4 as shown, Figure 1~4 The sensitivity detection results of the detection primers and probes of Bacteroides polymorpha, Clostridium tenabilis, Clostridium sphaericus and Lachnospira are shown in turn. It can be seen from the figure that the detection r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com