Salmonella and bacteria total number rapid synchronous multiple detection method and kit
A technology for Salmonella and total bacterial counts, applied in biochemical equipment and methods, microbiological-based methods, measuring devices, etc., can solve problems such as difficult multiple detection of Salmonella and total bacterial counts, inability to identify all types of bacteria, time-consuming and labor-intensive problems, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] The preparation of embodiment 1 Salmonella antibody
[0085] 1. Materials and methods
[0086] 1. A total of 2 experimental rabbits, with an initial weight of 2.3-2.5kg. Immunization method: multiple intradermal injections on the back. Immunogen: Salmonella-specific membrane protein. Process: A total of three immunizations were carried out with an interval of half a month. Then carry out the potency test. Then the fourth immunization was carried out, and the serum was collected after the titer test was suitable, and the antibody was purified. Got 2 Salmonella antibodies.
[0087] 2. Use the ELISA endpoint method to detect the titer of the obtained 2 Salmonella antibodies. Antibody concentration was detected by SDS-PAGE method.
[0088] 2. Experimental results
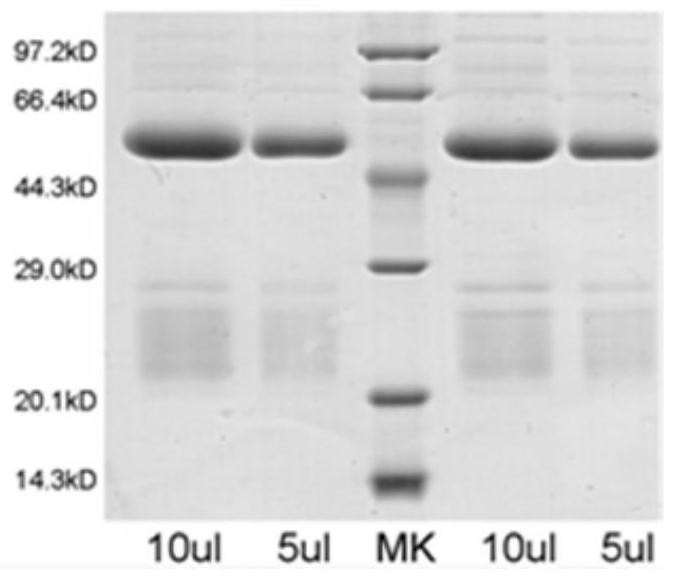
[0089] Preliminary preparation of 2 Salmonella, antibody 1 titer 1:1.25×10 6 , antibody 2 titer 1:3.125×10 5 . The electrophoresis results are as follows figure 1 shown.
[0090] Table 1 Antibody Tit...
Embodiment 2
[0095] Evaluation of Example 2 Salmonella Antibody
[0096] 1. Materials and methods
[0097] 1. Use the FITC labeling kit to label the prepared 8 Salmonella antibodies (Ab-1 to Ab-8) with fluorescein FITC.
[0098] 2. Salmonella paratyphi A, Salmonella typhimurium, Salmonella choleraesuis, Salmonella bonarene, Salmonella Kentucky, Salmonella enteritidis, Salmonella Sanftenburg, Salmonella abatetus. were prepared at a concentration of approximately 1×10 7 cells / mL of the above-mentioned Salmonella bacterium liquid.
[0099] 3. Add FITC-labeled antibodies (Ab-1 to Ab-8) at a concentration of 1 μg / mL to the bacterial liquids of the above eight strains of Salmonella, and incubate for 15 minutes in the dark. They were detected by fluorescence microscope and flow cytometer respectively.
[0100] 2. Experimental results
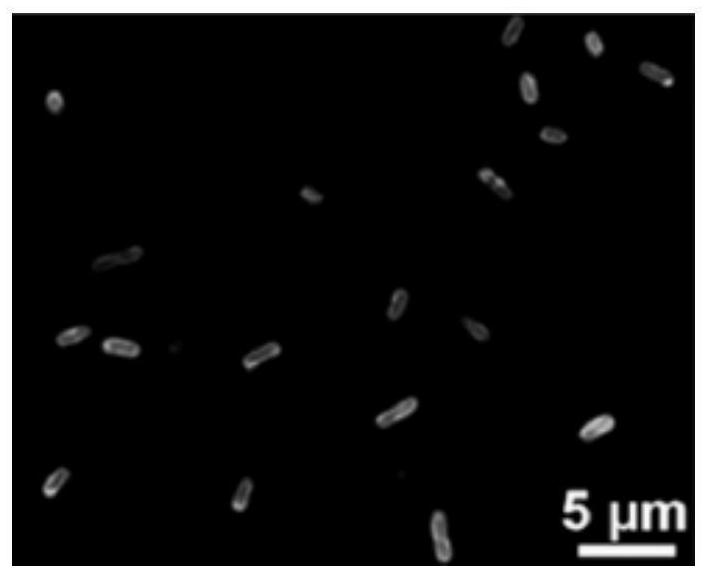
[0101] When labeled with Ab-1, Ab-4, Ab-6, and Ab-7, the eight strains of Salmonella all detected green fluorescence ( figure 2 ). When labeled with Ab-2 a...
Embodiment 3
[0106] Example 3 Affinity Analysis of Salmonella Antibody and Salmonella Membrane Protein
[0107] 1. Materials and methods
[0108] 1. Salmonella paratyphi A, Salmonella typhimurium, Salmonella choleraesuis, Salmonella bonarene, Salmonella Kentucky, Salmonella enteritidis, Salmonella Sanftenburg, Salmonella abatetus.
[0109] 2. Dilute the 4 Salmonella antibodies (Ab-1, Ab-4, Ab-6, Ab-7) obtained from the preliminary screening and 5 commercially purchased Salmonella antibodies (No. Ab-9 to Ab-13) to the same initial concentration, and 2-fold serial dilutions were performed.
[0110] 3. Separately prepare a concentration of about 1×10 7 Cells / mL of the above-mentioned Salmonella bacterium liquid was killed by heating to lyse its cell membrane, and then use the antibody to the structural antigen of Salmonella to carry out the fishing reaction to capture the specific membrane protein of Salmonella.
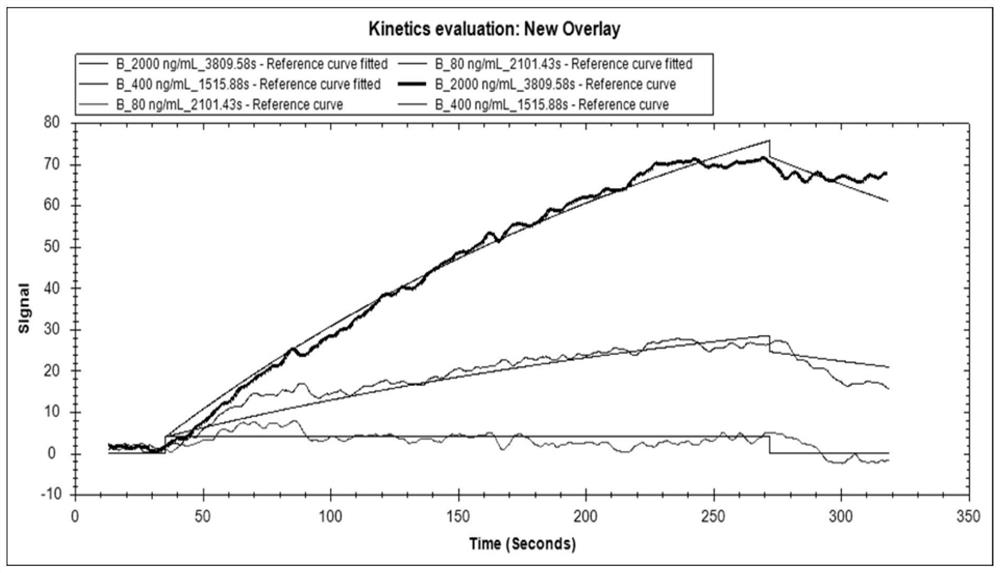
[0111] 4. Using the EDC / NHS reaction, link Salmonella-specific membrane prot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com