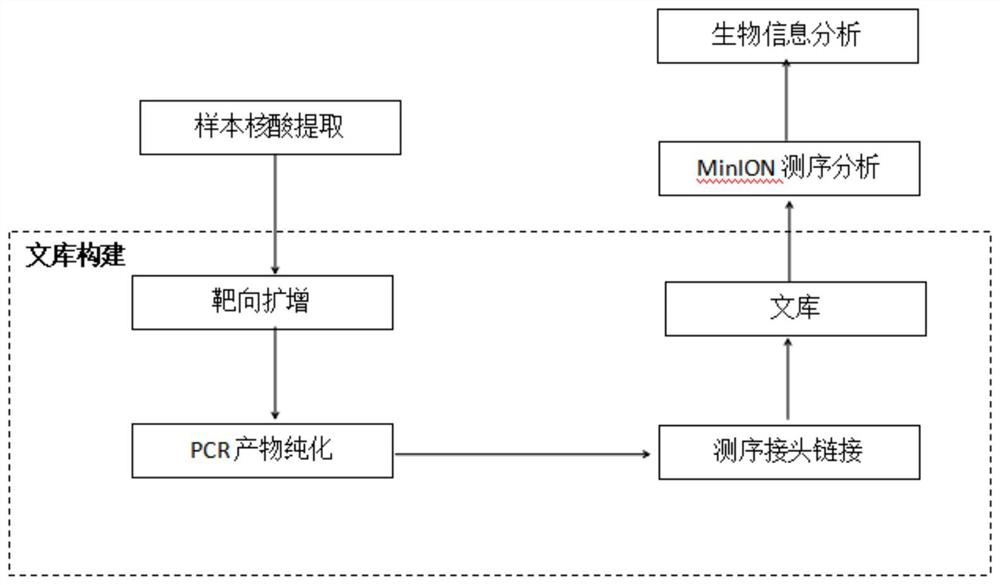
Primer group and kit for rapidly identifying respiratory tract microorganisms based on nanopore sequencing and application of primer group
A nanopore sequencing and kit technology, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, recombinant DNA technology, etc., and can solve problems such as sequence analysis that cannot be pathogenic
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] The design of embodiment 1 respiratory tract microorganism detection primer
[0083] According to bibliographical reports and epidemiological trend, the present embodiment designs the antimicrobial agent for 14 kinds of respiratory microorganisms (Streptococcus pneumoniae, Staphylococcus aureus (methicillin resistance), Klebsiella pneumoniae, Pseudomonas aeruginosa, Baumannii Detection of Acinetobacter, Stenotrophomonas maltophilia, Candida albicans, Haemophilus influenzae, Legionella pneumophila, Enterococcus faecium, Chlamydia psittaci, Cryptococcus gattii, Aspergillus fumigatus, Pneumocystis jirovecii) primers.
[0084] The detection objects designed in this embodiment include common microorganisms such as bacteria, fungi, and viruses. The above-mentioned microorganisms were identified by designing different target regions, among which (1) bacteria: 4 pairs of primers were designed to amplify the V1-V3, V4-V5, V6-V7 and V8 regions of 16s rRNA respectively, and the s...
Embodiment 3
[0088] Embodiment 3 Respiratory tract microorganism detection kit and its application method
[0089] 1. Composition
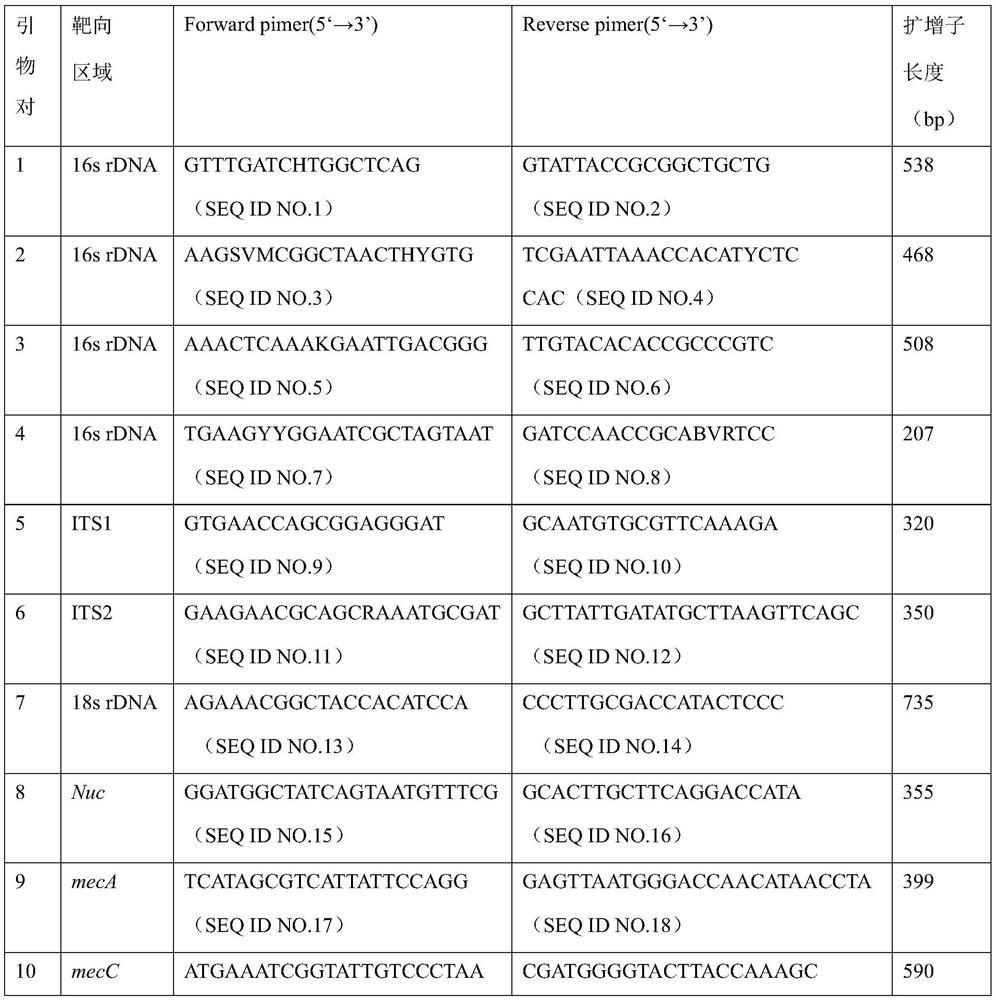
[0090]The nucleotide sequence is as shown in the primer set of SEQ ID NO: 1-20, and the primer set is made into multiple PCR primer mixture, and the concentration of each primer is 400nM-1μM, and the concentration of each primer is shown in Table 3;
[0091] table 3:
[0092]
[0093]
[0094] Library construction reagents: multiplex PCR amplification enzyme, ligation reaction mixture, eluent, positive quality control, negative quality control and internal standard, wherein the positive quality control is the target amplified fragment containing the primer set Cloning plasmid, negative quality control product is human genomic DNA, internal standard is MS2 phage DNA, multiplex PCR amplification enzyme contains Taq enzyme (0.5U / μl) and dNTPs (0.8mM), ligation reaction mixture is DNA ligase (1U / μl) μl) and buffer mixture, the eluent is 1×TE;
[0095] Lin...
Embodiment 4
[0134] The detection of embodiment 4 respiratory tract microbial samples
[0135] Collection of Streptococcus pneumoniae, Staphylococcus aureus (methicillin resistant), Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Stenotrophomonas maltophilia, Candida albicans, Haemophilus influenzae Bacillus, Legionella pneumophila, Enterococcus faecium, Chlamydia psittaci, Cryptococcus gattii, Aspergillus fumigatus, Pneumocystis jirovecii each 1 case; use the kit of Example 2 to detect.
[0136] The results are shown in Table 5, all of which were determined to be corresponding microorganisms.
[0137] table 5:
[0138] Numbering pathogen number of reads 1 Streptococcus pneumoniae (5) 2 Methicillin-resistant Staphylococcus (7) 3 Klebsiella pneumoniae (2) 4 Pseudomonas aeruginosa (1) 5 Acinetobacter baumannii (2) 6 Stenotrophomonas maltophilia (3) 7 Candida albicans (1) 8 Haemophilus influenzae (1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com