Method for detecting adsorption completeness of recombinant novel coronavirus vaccine
A coronavirus and detection method technology, applied in the field of recombinant novel coronavirus vaccine adsorption completeness detection, to achieve the effect of accurate and reliable experimental results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
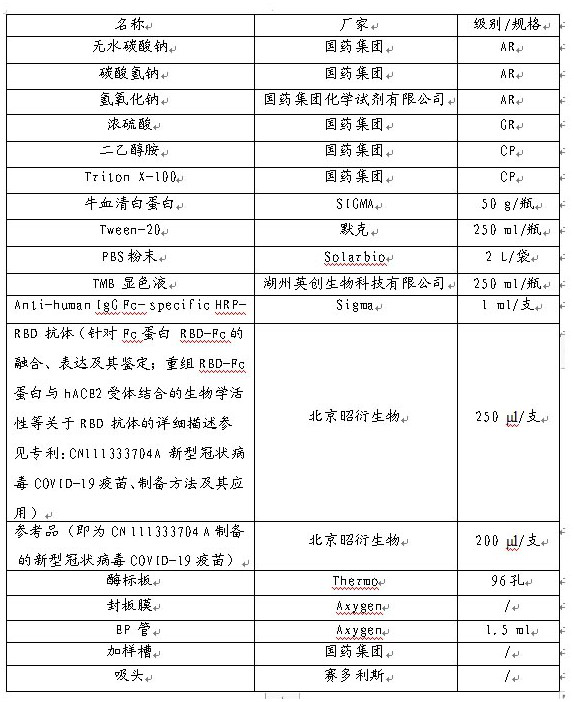
[0048] Solution preparation:
[0049] Coating solution: weigh 2.93 g NaHCO 3 , 1.59 g Na 2 CO 3 Dissolve in 800 ml, adjust the pH to 9.5-9.6 with 1M NaOH, and dilute to 1000 ml.
[0050] Blocking solution: Weigh 2.0 g of bovine serum albumin, dissolve in 100 ml of PBS solution, mix well and set aside.
[0051] Stop solution: Measure 42 ml of concentrated sulfuric acid, slowly add to 400 ml of water, stir while adding, and dilute to 750 ml after cooling.
[0052] Sample Diluent / Wash: PBS with 0.05% Tween-20.
[0053] Test product / reference product treatment solution: Measure 1.25 ml of 20% diethanolamine and 0.20 ml of 10% Triton X-100, add 8.55 ml of PBS, mix well and set aside.
[0054] Reference and test product handling:
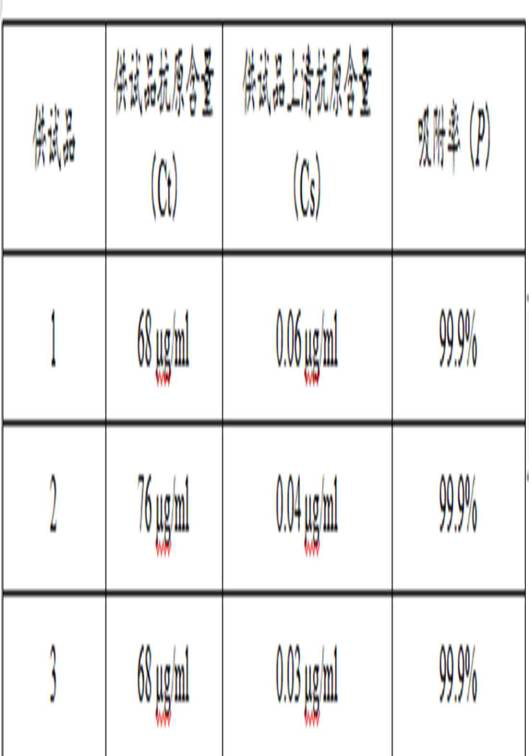
[0055] Desorption of the test product: Precisely measure 0.2 ml of the same batch of test product 1, test product 2 and test product 3 and place them in three 1.5ml EP tubes, add 0.2 ml of test product / reference product respectively for treatment ...
Embodiment 2
[0061] A method for detecting the completeness of recombinant novel coronavirus vaccine adsorption, comprising the following steps:
[0062] Coating: After diluting the RBD antibody with a concentration of 2.1 mg / ml to 5 μg / ml, pre-coat it into a 96-well microtiter plate with a coating volume of 100 μl / well, and place it at 6°C for 15 hours.
[0063] Sealing: Take the coated plate out from 2-8°C, wash the plate 4 times, each washing volume is 300 μl / well, pat dry on absorbent paper, add blocking solution, 300 μl / well, cover the plate membrane , and incubate at 37±1°C for 90 minutes.
[0064] Adding samples: Take out the closed ELISA plate, add 300 μl / well of washing solution, shake gently for about 30 seconds, discard the washing solution, pat dry on absorbent paper, and repeat washing 4 times; dilute the solution prepared in Example 1 well The working reference substance, the quality control substance, the desorption diluent of the test product, and the undesorbed supernatan...
Embodiment 3
[0070] data processing:
[0071] 1) Standard curve drawing: take the concentration value of the reference product as the abscissa (ng / ml), and the corresponding average OD value after deducting the negative control absorbance (OD) value as the ordinate, perform four-parameter fitting to generate the S curve and If the four-parameter equation, standard curve and quality control data meet the criteria for system applicability, the fitting is passed;
[0072] Fitting curve R 2 If it is less than 0.990, the experiment should be repeated.
[0073] The resulting standard curve equation is as follows:
[0074] y = (A - D) / [1 + (x / C) B ] + D type c;
[0075] In the formula, A= 2.00742; B = -0.82100; C= 314.08530; D= -0.09311.
[0076] R 2 It is 0.99981, which meets the requirements.
[0077] 2) Calculation of the recovery rate of quality control products: Substitute the absorbance values of QCH and QCL after subtracting the OD value of the negative control into the standard...
PUM
| Property | Measurement | Unit |
|---|---|---|
| adsorption capacity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com