Molecular beacon probe and kit for detecting equine influenza virus pathogens
A technology of molecular beacon probes and kits, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of difficult identification, small number of samples, and slow detection speed, etc. Effects of background ratio, improved repeatability and accuracy, and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1) Design primers and molecular beacon probes
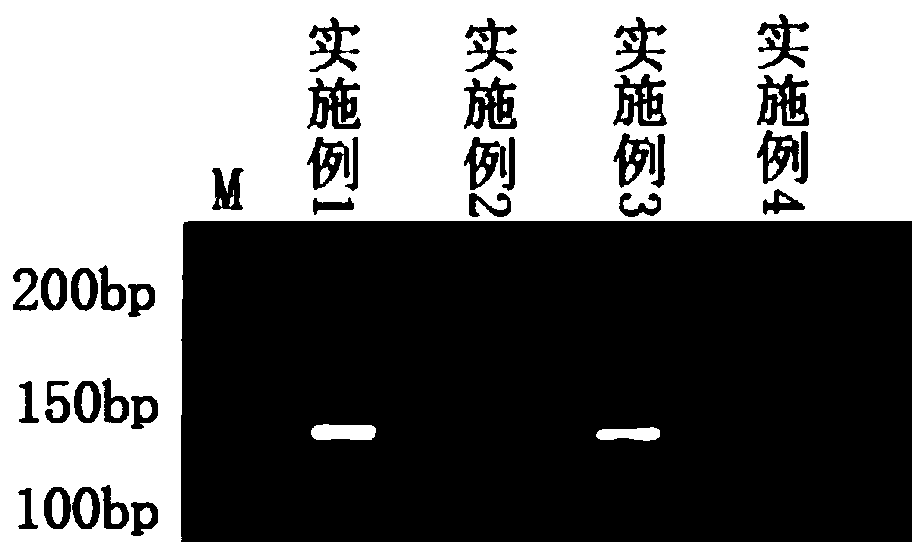
[0041]Design primers and molecular beacon probes according to the partial nucleotide sequence of the PB1 gene of H3N8 downloaded from the GenBank database, wherein the partial nucleotide sequence of the PB1 gene is a 145bp fragment:
[0042] AAGGCCGGCAAACTTATGATTGGACCTTGAATAGGAATCAACCTGCTGCAACAGCACTTGCTAATACAATTGAAGTGTTCAGATCAAATGGTCTGACTTCCAATGAATCAGGGAGATTGATGGACTTCCTCAAAGATGTCATGGA
[0043] The primer pairs thus synthesized are:
[0044] PF:AGAATAAGTAGACAATCCAT;
[0045] PR: GGTATATGCTTCAGTACGTACA;
[0046] Molecular beacon probes are:
[0047] P probe
[0048] 5'Fam-CCGACAACTTATGATTGGACCTTGAAGTCGG-Dabcyle 3', the molecular beacon is a specific sequence of a neck ring structure composed of bases, in which the 5' end is marked with FAM, and the 3' end is marked with Dabcyle, the excitation wavelength of the fluorophore It is 494nm, and the emission wavelength is 515nm.
[0049] 2) Construction and preparation of qu...
Embodiment 2
[0070] 1) Design primers and molecular beacon probes
[0071] According to the nucleotide sequence of the HA gene of H3N8 downloaded from the GenBank database, select a highly conserved and specific nucleotide sequence to design primers and molecular beacon probes. The highly conserved and specific nucleotide sequence of the HA gene For a 172bp fragment:
[0072] ACCCATATGACATCCCTGACTATGCATCGCTCCGATCAATTGTAGCATCCTCAGGAACATTGGAATTCACAGCAGAGGGATTCACATGGACAGGTGTCACTCAAAACGGAAGAAGTGGAGCCTGCAAAAGGGGATCAACCGATAGTTTCTTTAGCCGACTGAATTGGCTAAC
[0073] The primer pairs thus synthesized are:
[0074] HF:GTACGTGGATCTCCTATGAC;
[0075] HR: CTTATGTGGCTCCTAATCTAGC;
[0076] Molecular beacon probes are:
[0077] H probe
[0078] 5'Fam-CCGACAACATTGGAATTCACAGCAGAGTCGG-Dabcyle 3', the molecular beacon is a specific sequence of a neck ring structure composed of bases, in which the 5' end is marked with FAM, and the 3' end is marked with Dabcyle, the excitation wavelength of the fluorophore I...
Embodiment 3
[0080] 5) Molecular beacon fluorescent quantitative PCR reaction:
[0081] With the cDNA obtained in step 4) as a template, the amplification system is as follows:
[0082] 2.5 μL 1.2mmol / L 10×PCR reaction buffer, 4 μL 15mmol / L MgCl 2 , 1 μL 50pmol / L upstream primer, 1 μL 50pmol / L downstream primer, 1 μL 50pmol / L molecular beacon probe, 2 μL 2mmol / L dNTP, 0.5U Taq enzyme, 1 μL DNA template, 1 μL 2.5mmol / L laconic acid, ddH 2 O supplemented to 25 μL. The reaction conditions for PCR amplification are: denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, amplification at 72°C for 30 seconds, and 40 PCR cycles; Fluorescent signal was detected. All the rest are completely consistent with Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com