Molecular conformation searching method based on hybrid fireworks algorithm
A firework algorithm and molecular conformation technology, applied in the field of computer-aided drug design, can solve problems such as low prediction accuracy and long time for molecular docking, and achieve the effects of improving accuracy, reducing average time, and increasing speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
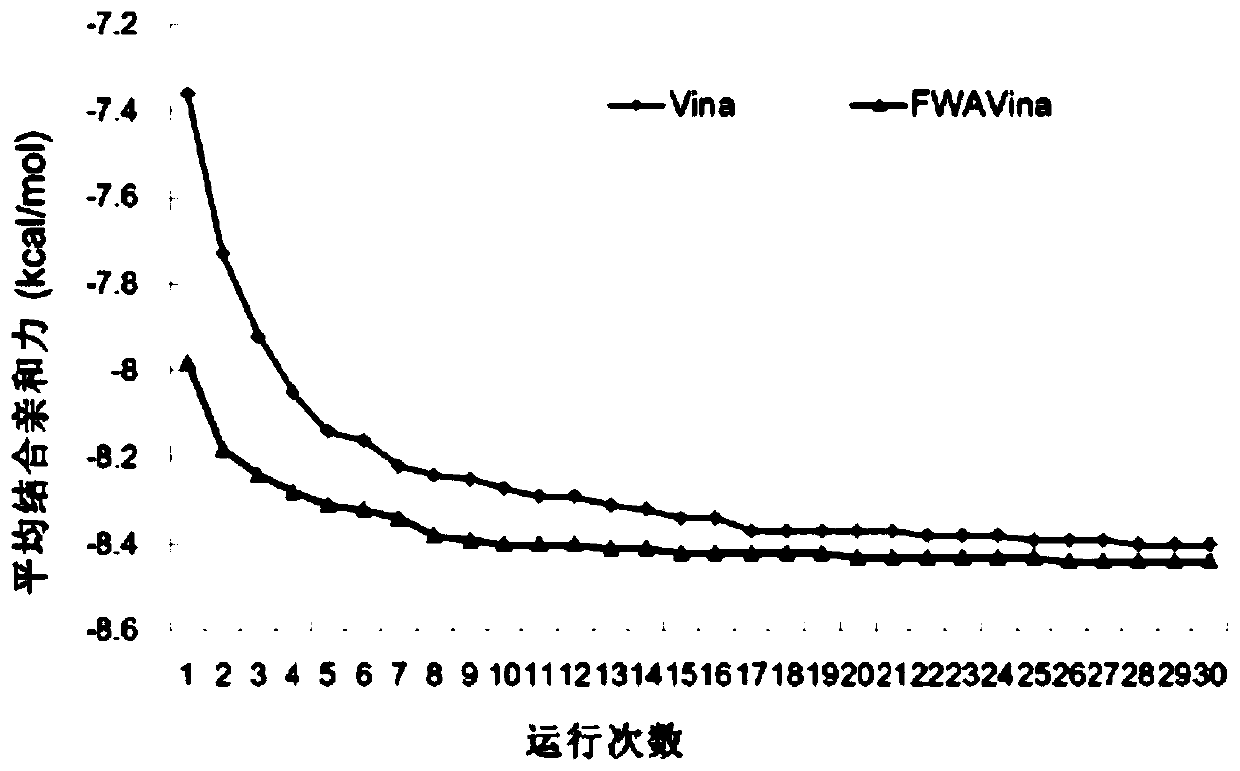
[0055] In Example 1, the docking of 195 receptor-ligand complexes in the core set of the PDBbind standard test set was used as the test object, and the search method of the present invention was used for molecular docking, and each complex docking was run 30 times. figure 2 It is a flow chart of Embodiment 1 of the present invention, combined below figure 2 to elaborate.
[0056] Preferably, this embodiment is implemented in the framework of the molecular docking software Autodock Vina (abbreviated as Vina), and the scoring function of Vina is used as the fitness function. Specific steps are as follows:
[0057] (1) Convert the pdb file of the protein and the mol2 file of the ligand into pdbqt files respectively;
[0058] (2) Set the range of the receptor binding pocket, that is, set the center position of the docking box (center_x, center_y, center_z), length, width, and height (size_x, size_y, size_z);
[0059] (3) Generate initial fireworks: initialize N initial firewo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com