Method for preparing (S)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid through chemical enzyme process
A technology of tetrahydroisoquinoline and chemical enzymatic method, applied in the field of biocatalysis, can solve problems such as the theoretical yield of only 50%, and achieve the effects of high reaction efficiency and yield, mild reaction conditions and improved yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1D
[0056] Example 1 Screening of D-amino acid oxidase and construction of genetically engineered bacteria expressing D-amino acid oxidase
[0057] According to different substrate specificities, D-amino acid oxidases from microorganisms can be divided into two categories: 1) amino acids (such as D-alanine) with a preference for substrate side chain groups, such as Fusarium oxysporum (Fusarium oxysporum)-derived D-amino acid oxidase; 2) preference for amino acids with larger substrate side chain groups (such as D-phenylalanine), such as Trigonopsis variabilis-derived D-amino acid oxidase ( POLLEGIONI L, MOLLAG, SACCHI S, et al. Properties and applications of microbial D-amino acid oxidases: current state and perspectives [J]. Appl Microbiol Biotechnol, 2008, 78(1): 1-16.). The amino acid sequences of these two D-amino acid oxidases were used to perform BLASTp analysis in the National Center for Biotechnology Information (NCBI) database (https: / / www.ncbi.nlm.nih.gov / ), and the sequ...
Embodiment 2
[0062] 2.1 Culture of microorganisms
[0063] Composition of liquid LB medium: peptone 10g / L, yeast powder 5g / L, NaCl 10g / L, dissolved in deionized water and then constant volume, sterilized at 121°C for 20min, ready for use. If it is solid LB medium, add 15g / L agar.
[0064] The engineered bacteria containing the D-amino acid oxidase gene were inoculated in 5 mL of liquid LB (containing 50 μg / ml kanamycin) medium, and cultured with shaking at 200 rpm for about 8 hours at 37°C. Inoculate in 100mL liquid LB (containing 50μg / ml kanamycin) culture medium with 1% (V / V) inoculum size, OD 600 After reaching 0.6-0.8, add the inducer isopropylthiogalactopyranoside (final concentration: 0.1 mM), and induce for 15 hours at 18°C. After the cultivation, pour the culture solution into a 100mL centrifuge tube and centrifuge at 4000rpm for 10min, discard the supernatant, collect the bacterial cells, wash the cells twice with 50mM phosphate buffer (pH=8.0), and store them in a -80°C ultra-l...
Embodiment 3
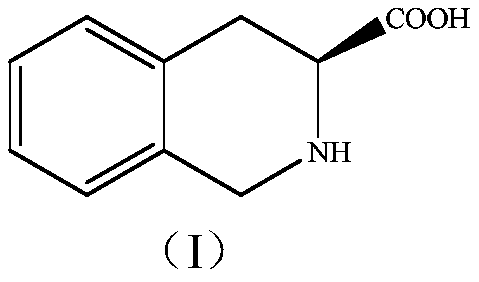
[0070] Example 3 FsDAAO-NH 3 ·BH 3 Preparation of (S)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid
[0071] Preparation of substrate solution: Prepare 5 g / L racemic 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid solution with 50 mM phosphate buffer solution (pH=8.0) and adjust the pH of the solution with 30% ammonia water value to 8.0.
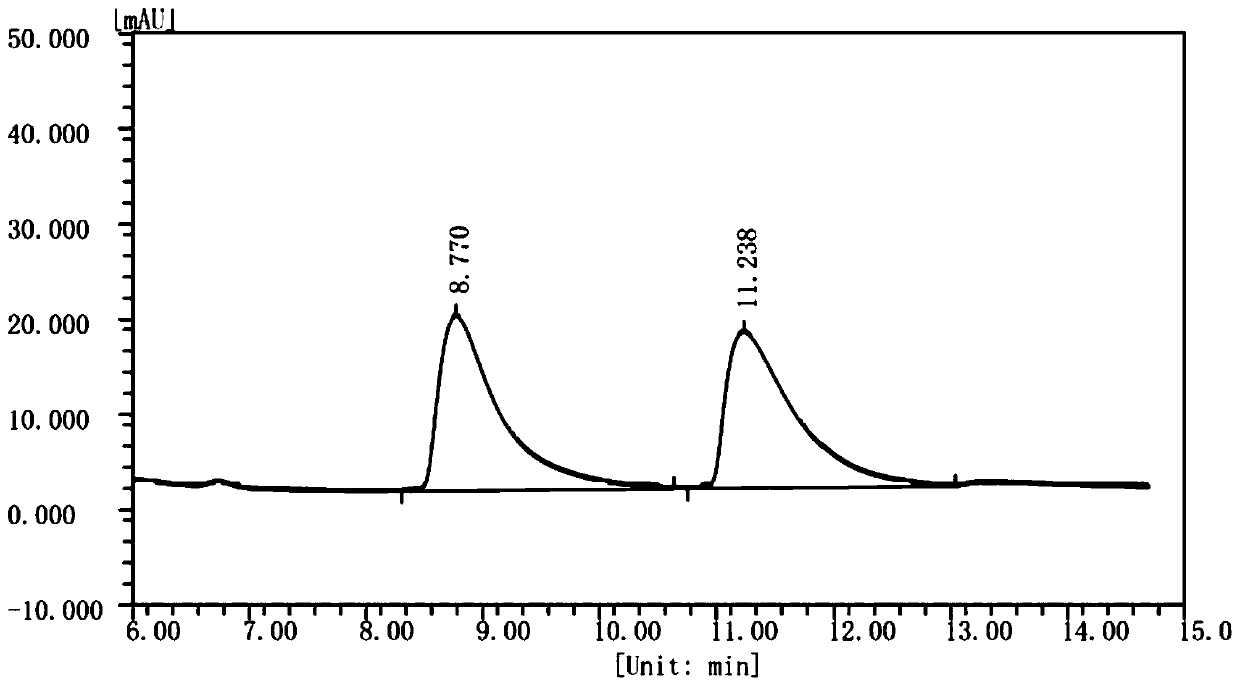
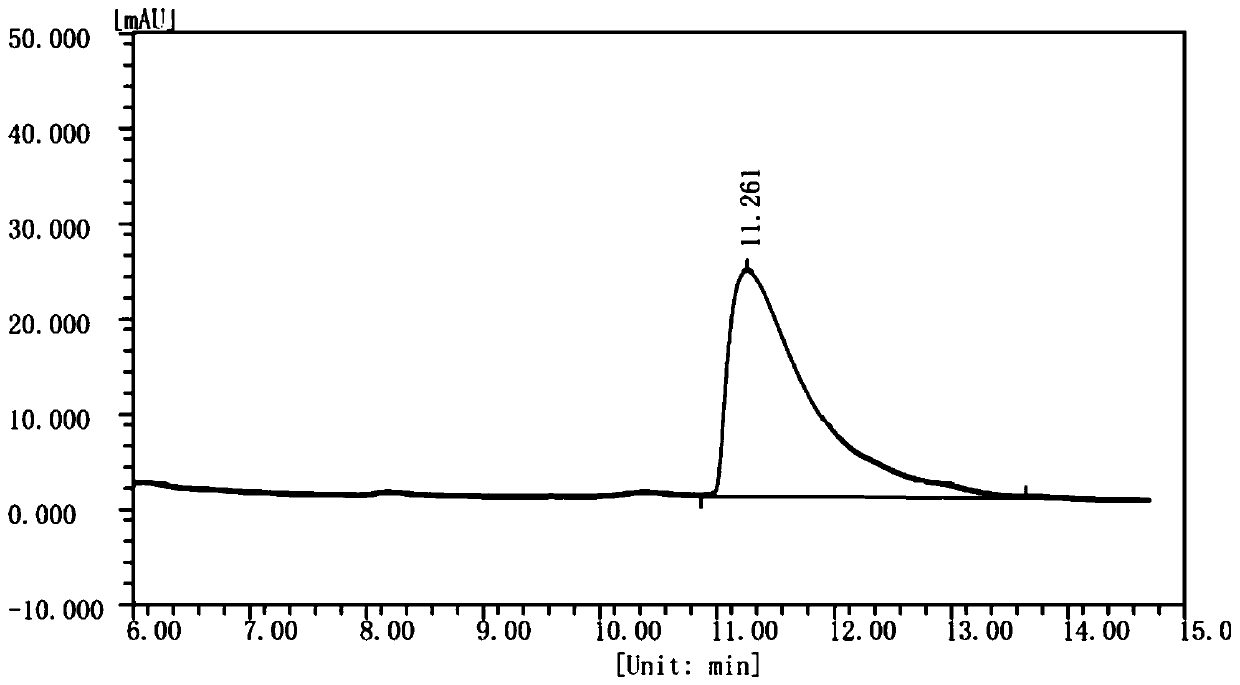
[0072] Add 24mL substrate solution, 6mL FsDAAO crude enzyme solution (the crude enzyme solution already contains enough coenzyme FAD, therefore, no additional FAD needs to be added in the crude enzyme solution reaction system), 12mg catalase lyophilized to 100mL reactor powder and 0.4gNH 3 ·BH 3 . Immediately after mixing, a sample was taken as "0 hour". The reaction system was placed in a constant temperature water bath at 30° C., stirred by magnetic force, and reacted for 24 hours before sampling. The content of two configurations of 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid in the sample was detected by high performance ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| enantiomeric excess | aaaaa | aaaaa |
| enantiomeric excess | aaaaa | aaaaa |
| enantiomeric excess | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com