Method for detecting number of soil bacteria based on real-time fluorescent quantitative PCR
A real-time fluorescent quantitative and bacterial technology, applied in the biological field, can solve the problems such as the inability to obtain reliable information on the number of bacteria by the dilution plate method, the inappropriate promotion of high-throughput sequencing, and the low accuracy of observation and counting. Difficulty, easy to operate and easy to learn
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
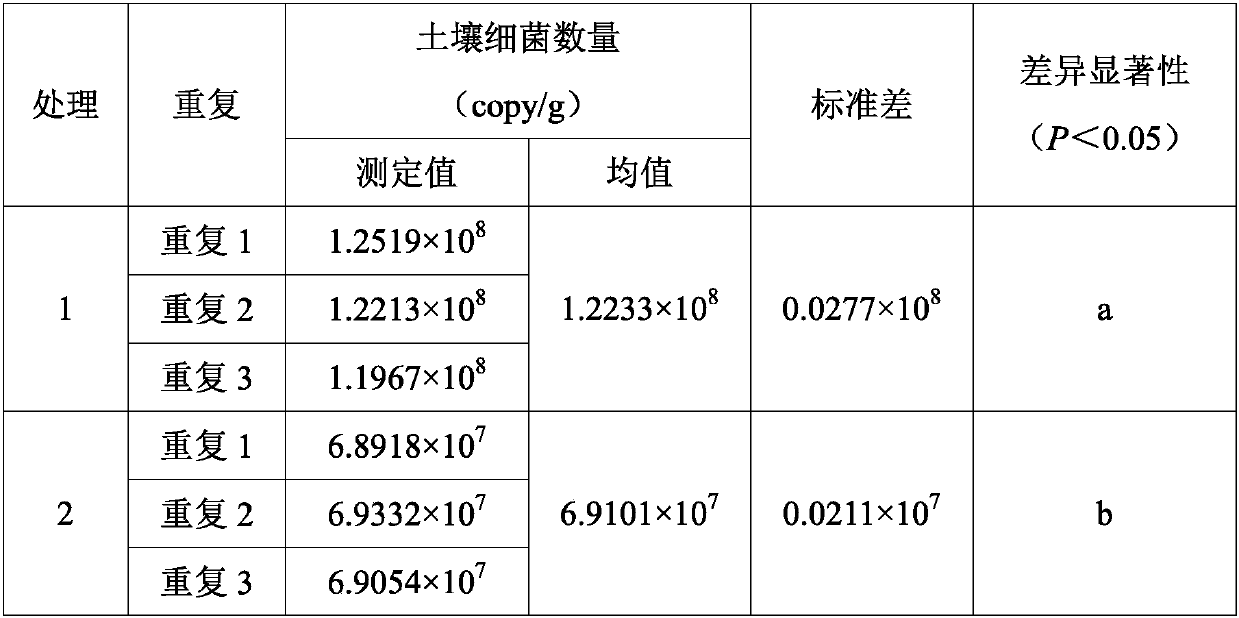
[0027] The soil used in this example was collected from fertilized bamboo forests and unfertilized bamboo forests at the northern foot of Zijin Mountain (32°01'N, 118°49'E) in Nanjing City, Jiangsu Province. The soil type is yellow brown soil and weakly acidic. Remove the litter layer about 0.5m away from the trunk of the tree, and use a sterilized 50mL centrifuge tube to take the surface soil (0-10cm). Treatment 1 is the fertilized bamboo forest soil, and treatment 2 is the non-fertilized bamboo forest soil. Each treatment is repeated 3 times. Methods To detect the number of soil bacteria.
[0028] The specific operation steps are as follows:
[0029] 1) Pass the soil sample through a 2mm sieve to remove debris such as stones, tree roots and fallen leaves, weigh about 0.3g of fresh soil, and use the OMEGA Soil DNA Extraction Kit (OMEGA E.Z.N.A. Soil DNA Extraction Kit type is 50 times / box , Cat. No. D5625-01) to extract total DNA from soil, dissolved in ddH 2 In O, the DNA ...
Embodiment 2
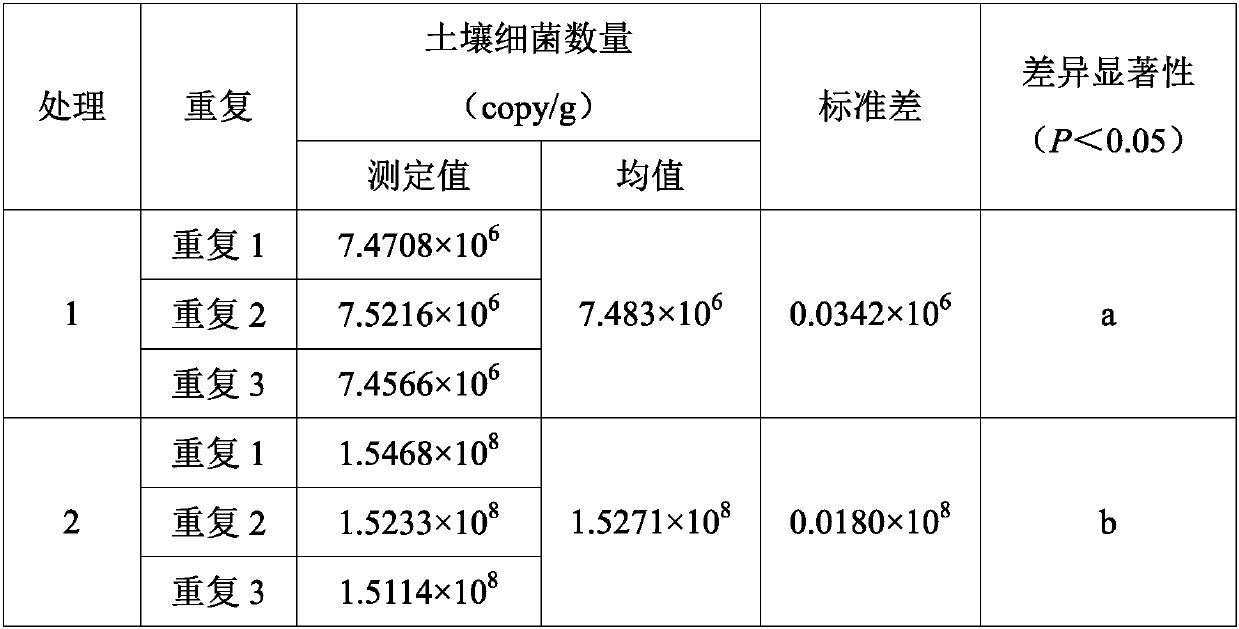
[0044] The soil used in this example was collected from the South Seedling Base (21°27'N, 110°11'E) of Zhanjiang City, Guangdong Province, 4-year-old and 14-year-old Eucalyptus rugosa plantations. The soil type is brick red soil, which is weakly acidic. Remove the litter layer about 1m away from the main trunk of the tree, and take the surface soil (0-10cm) with a sterilized 50mL centrifuge tube. Treatment 1 is the soil of the 4-year-old Eucalyptus ivory plantation, and Treatment 2 is the soil of the 14-year-old Eucalyptus ivory plantation. 3 repetitions. Concrete operation step is with embodiment 1, draws this standard curve, y=-3.4484x+36.632; Wherein R 2 =0.9986, amplification efficiency E=94.98%. The results are shown in Table 2:
[0045] Table 2 Detection of soil bacteria in 4-year-old and 14-year-old Eucalyptus scutellaria plantations
[0046]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com