SgRNA sequence for targeted knockout of FcMYC2 gene, CRISPR/Cas9 vector and application thereof
A sequence and gene technology, applied in the field of sgRNA sequence targeting knockout of FcMYC2 gene, CRISPR/Cas9 vector
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] sgRNA oligonucleotide strand synthesis and annealing
[0049] Use the CRISPR-P 1.0 online design tool (http: / / crispr.hzau.edu.cn / CRISPR / ) to select and design two 20bp sgRNAs (FcMYC2sgRNA-1 and FcMYC2sgRNA-2) on FcMYC2 according to the scoring system, and pass BLAST verification free of non-specific genes. Add GATTG to the 5'-end of the positive strand of the sgRNA, add the AAAC sequence to the 5'-end of the complementary strand of the sgRNA, and add C to the 3'-end. The oligonucleotides shown in Table 1 were formed and synthesized by BGI.
[0050] Take out the synthetic sgRNA oligonucleotide sequence, add an appropriate amount of ddH2O to make a 100 μM solution, and prepare the annealing reaction solution according to the system in Table 2.
[0051] The prepared reaction solution was incubated at 95°C for 10 minutes, then removed, and buffered and cooled at room temperature. Store the annealed double-stranded oligonucleotides at -20°C for future use.
Embodiment 2
[0053] Construction of Knockout Citrus FcMYC2 Vector
[0054] The pPIC.5 and pPIC.1 used in this example were purchased from Jiangsu Jirui Biotechnology Co., Ltd.; the p1301NG vector used can be found in the literature nptII::mgfp5 fusion gene construction and its application in citrus genetic transformation (Liu Xiaofeng, Peng Aihong, Xu Lanzhen et al., Construction of nptⅡ::mgfp5 fusion gene and its application in citrus genetic transformation. Journal of Tropical Crops, No. 7, 2013).
[0055] The "CAMV35S+nptⅡ::mgfp5 fusion gene" was excised from the p1301NG vector with EcoRI / XhoI and ligated into pPIC.1 cut with the same restriction enzymes to obtain the NPTII selection marker and green fluorescent protein (GFP) reporter gene suitable for citrus transgenic use The expression vector pPICL (preserved in the laboratory, and promised to release to the public, can be used to repeat the experiment of the present invention).
[0056] Ligate the two annealed double-stranded oligo...
Embodiment 3
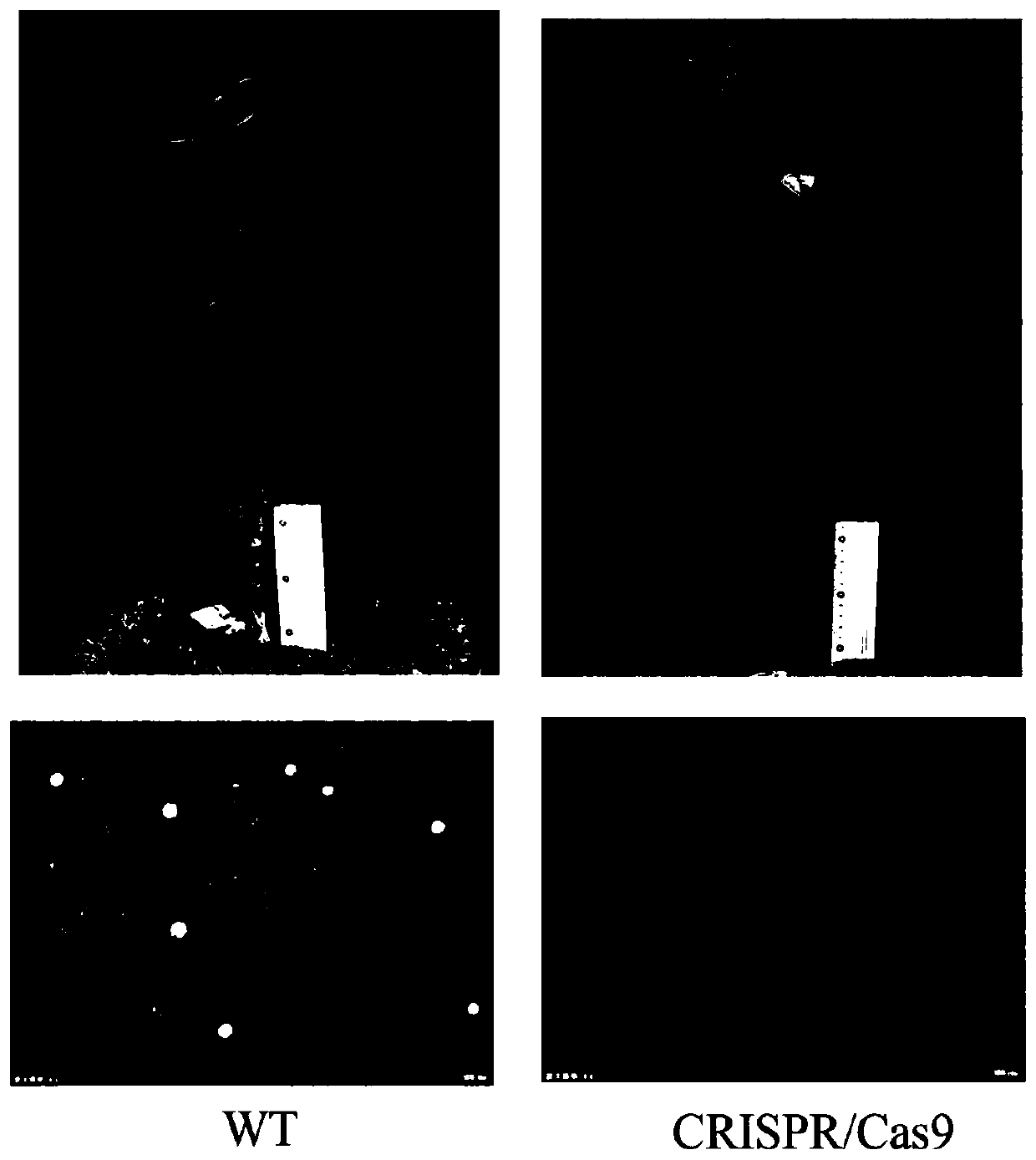
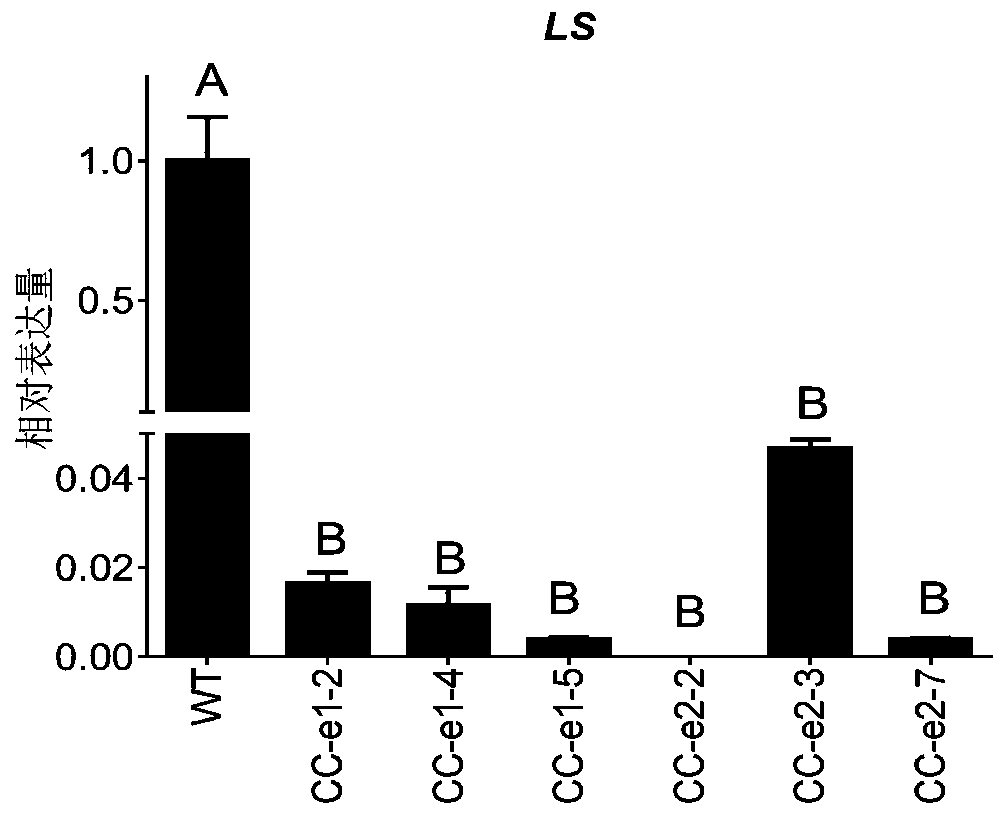
[0058] Obtaining Jincheng by knocking out FcMYC2 gene
[0059] The receptor material is the epicotyl of C. sinensis (L.) Osbeck cv. Jincheng. The model of MS basal medium is M519 (PhytoTechnology Laboratories, USA). All hormones and antibiotics are filter-sterilized and added to the medium at the time of use. The specific steps of genetic transformation are as follows:
[0060] (1) Cultivation of epicotyls
[0061] About 25 days before the transformation, take the ripe and undamaged Jincheng fruit, carefully wash it in warm water with detergent, dry it at room temperature, and soak it with 70% alcohol for 15 minutes on the ultra-clean workbench. Cut the fruit with a blade, take out the plump seeds, peel off the outer testa and inner testa, put into the germination medium [MS (5% Suc)+9g L -1 agarpowder, pH5.8] in a test tube, cultured in the dark at 28°C in an incubator. 3-5 days before the transformation, the test tubes will be taken out and cultured under light.
[006...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com