CRISPR-Cas9-based nucleic acid detection system and application thereof
A detection system and nucleic acid technology, applied in the fields of synthetic biology and biomedicine, can solve problems such as low reprogramming efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 Design standard sgRNA and RsgRNA
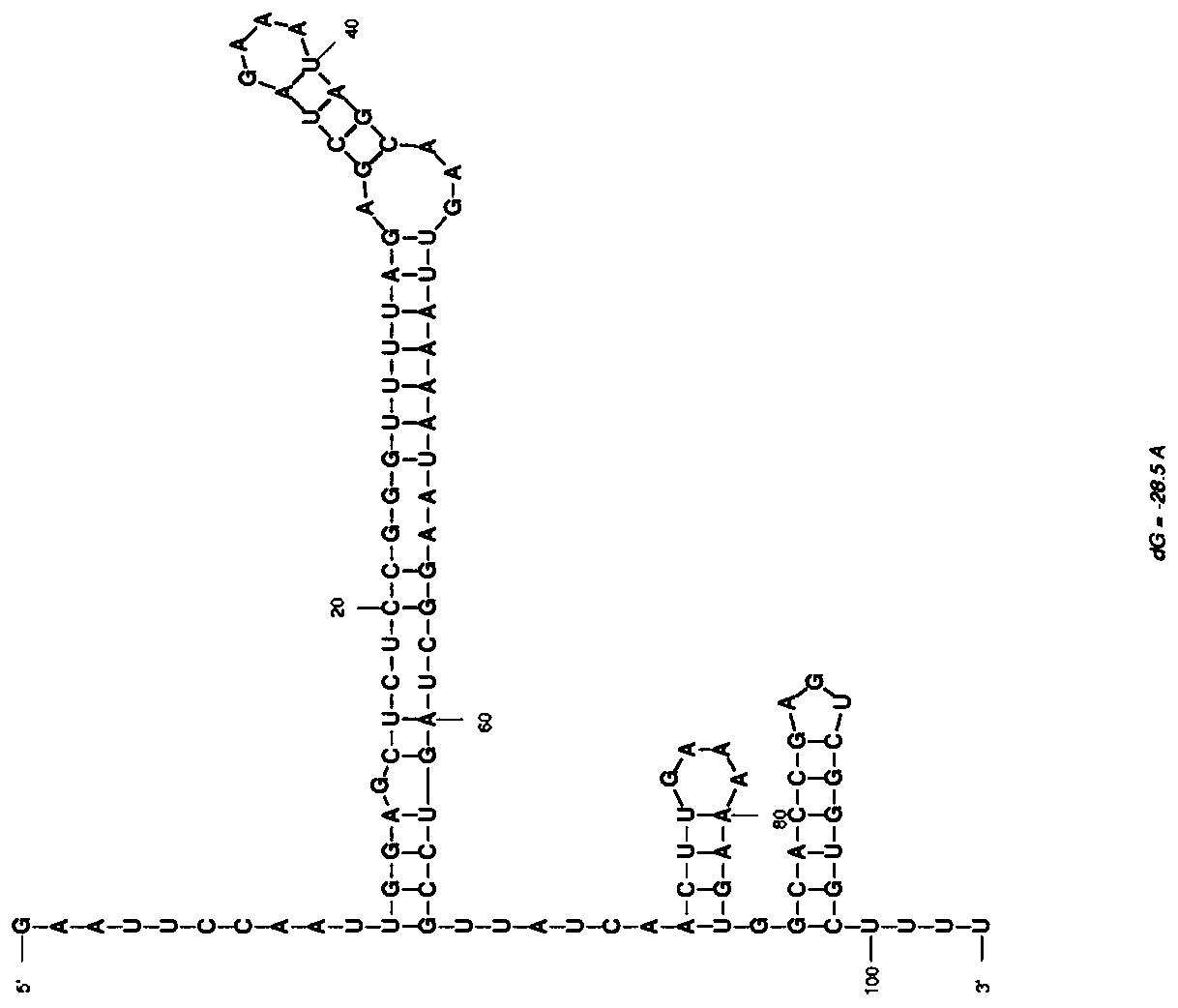
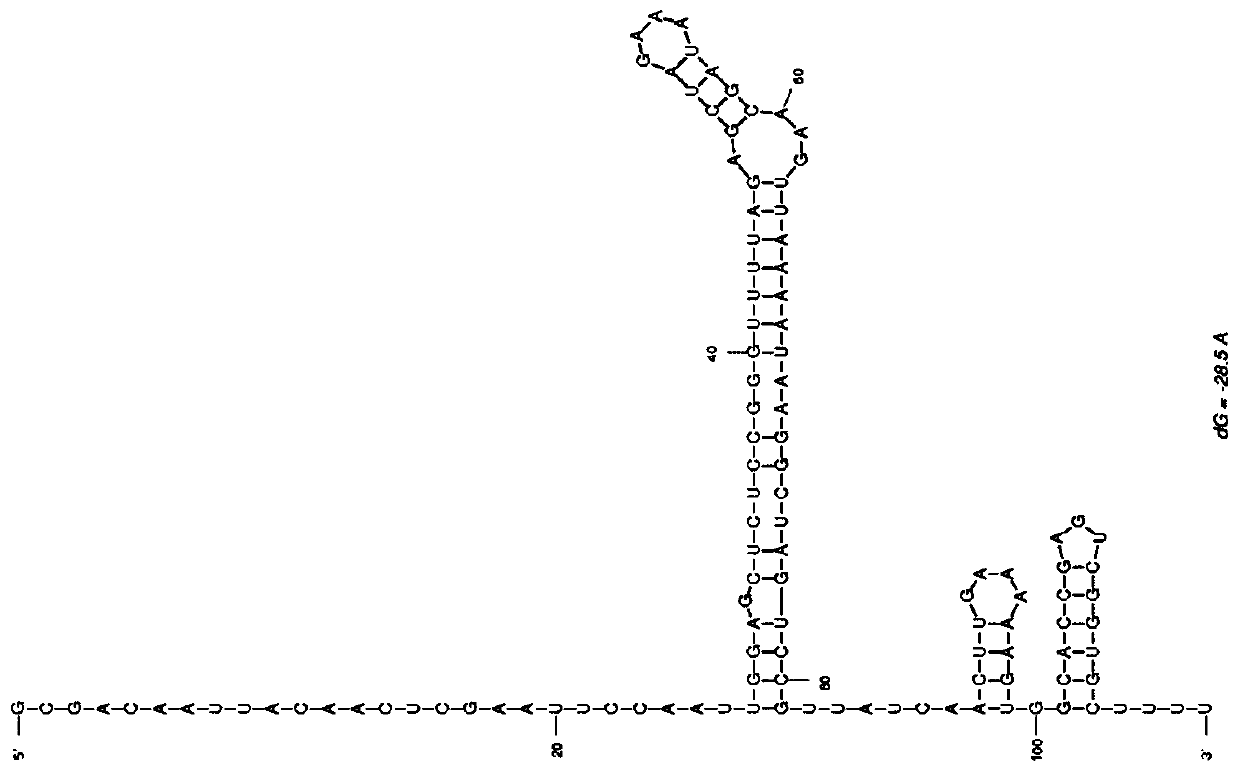
[0079] Standard sgRNA targets were designed by CHOPCHOP (https: / / chopchop.rc.fas.harvard.edu / ), and screened and evaluated by Moreno-Mateos scores (Vejnar C E, et al..Cold Spring Harbor Protocols, 2016), obtained as The sgRNA shown in SEQ ID NO:2, the secondary structure is as follows figure 1 Shown; The RsgRNA shown in SEQ ID NO:3 increases the antisense nucleic acid shown in SEQ ID NO:1 at the 5' end of sgRNA, and the secondary structure is as shown in figure 2 shown.
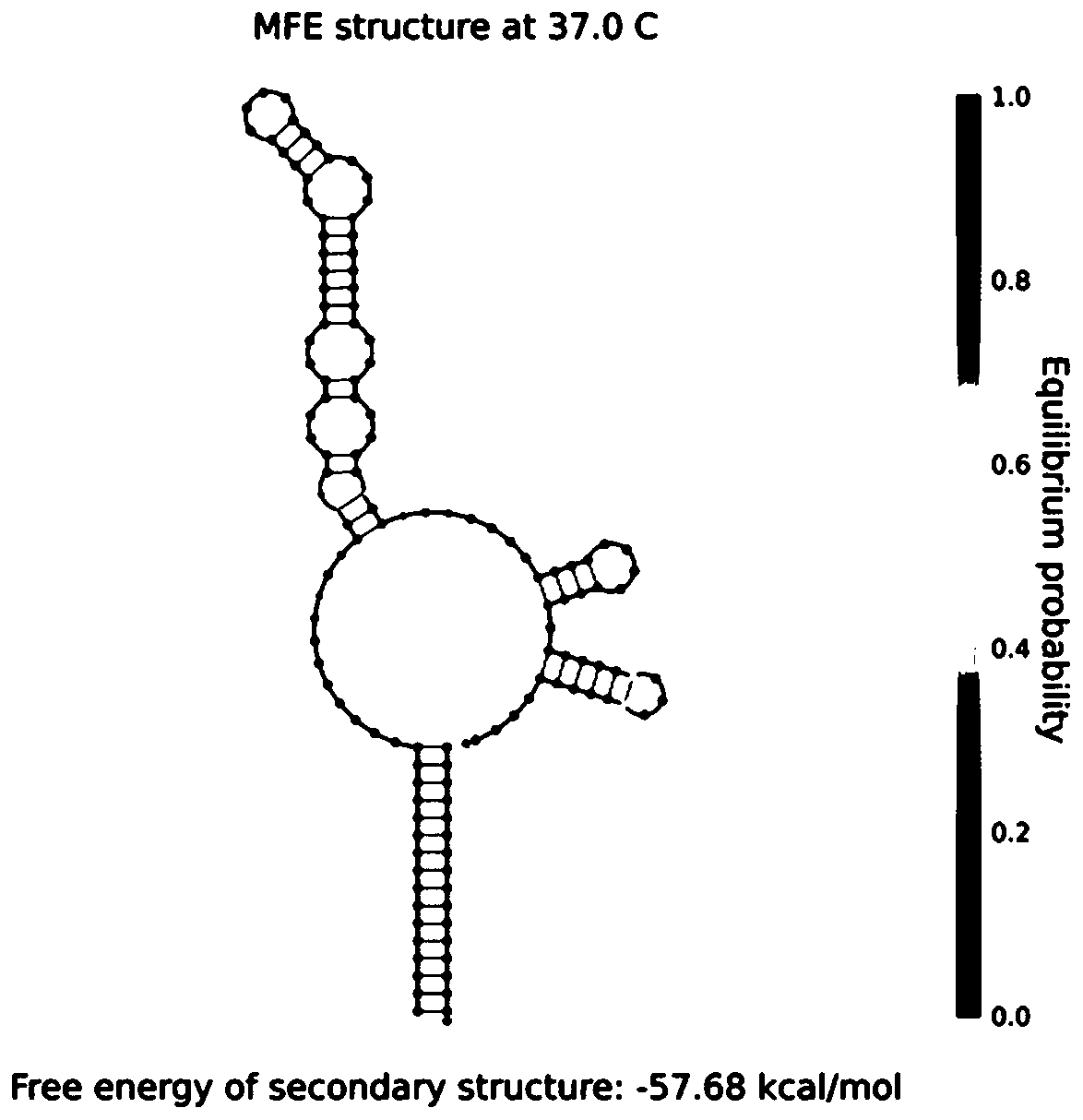
[0080] Such as image 3 As shown, in the presence of the target nucleic acid, the trigger sequence hybridizes with the antisense nucleic acid on the RsgRNA, and the free energy of the secondary structure ΔG=-57.68kcal / mol.
Embodiment 2
[0081] Construction of embodiment 2 reporter plasmid
[0082] The reporter plasmid constructed in this example includes T7 promoter, ribosome binding site, G6PD coding sequence and T7 terminator. According to the protein sequence of G6PD, combined with the characteristics of E. coli codon optimization, the design is shown in SEQ ID NO: 4 The G6PD coding sequence, the steps are as follows:
[0083] Using the Hanbio HB-infusionTM seamless cloning kit, using the DNA exonuclease, DNA polymerase and ligase in the system to insert the coding sequence shown in SEQ ID NO: 4 into the vector pET3a backbone, avoiding restriction internal Dicer cloning method on the impact of the promoter region and ribosome binding site sequence Shine-Dalgarno (SD), constructed as Figure 4 Plasmids indicated.
Embodiment 3
[0084] Example 3 Regulatory effect of Cas9-sgRNA or Cas9-RsgRNA on G6PD
[0085] The Cas9-sgRNA or Cas9-RsgRNA constructed in Example 1 was added to the cell-free protein expression system containing the reporter plasmid to regulate the synthesis of G6PD protein.
[0086] Such as Figure 5 Indicates the cleavage effect of Cas9-RsgRNA on DNA and the inhibitory effect of different concentrations of target substances on the cleavage substrate. Among them, the leftmost lane is the DNA ladder lane, lane 1 indicates DNA as the cleavage substrate, and lane 2 indicates Cas9-RsgRNA The cleavage products after cleavage of substrate DNA, lanes 3-8 represent the inhibitory effect of different concentrations of trigger sequences (0.1, 0.4, 2, 10, 50, 250nM) on the Cas9-RsgRNA cleavage system. The results showed that with the increase of the concentration of the trigger sequence, its inhibitory effect on Cas9-RsgRNA cleavage was stronger.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com