Identification method of symbiotic microorganisms of medicinal plants and application of identification method
An identification method and technology of medicinal plants, applied in the field of bioinformatics research, can solve the problems that the results are not necessarily scientific and practical, the analysis and identification period is long, and the accuracy is poor, and the identification period is short, the accuracy is high, and the time is short. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
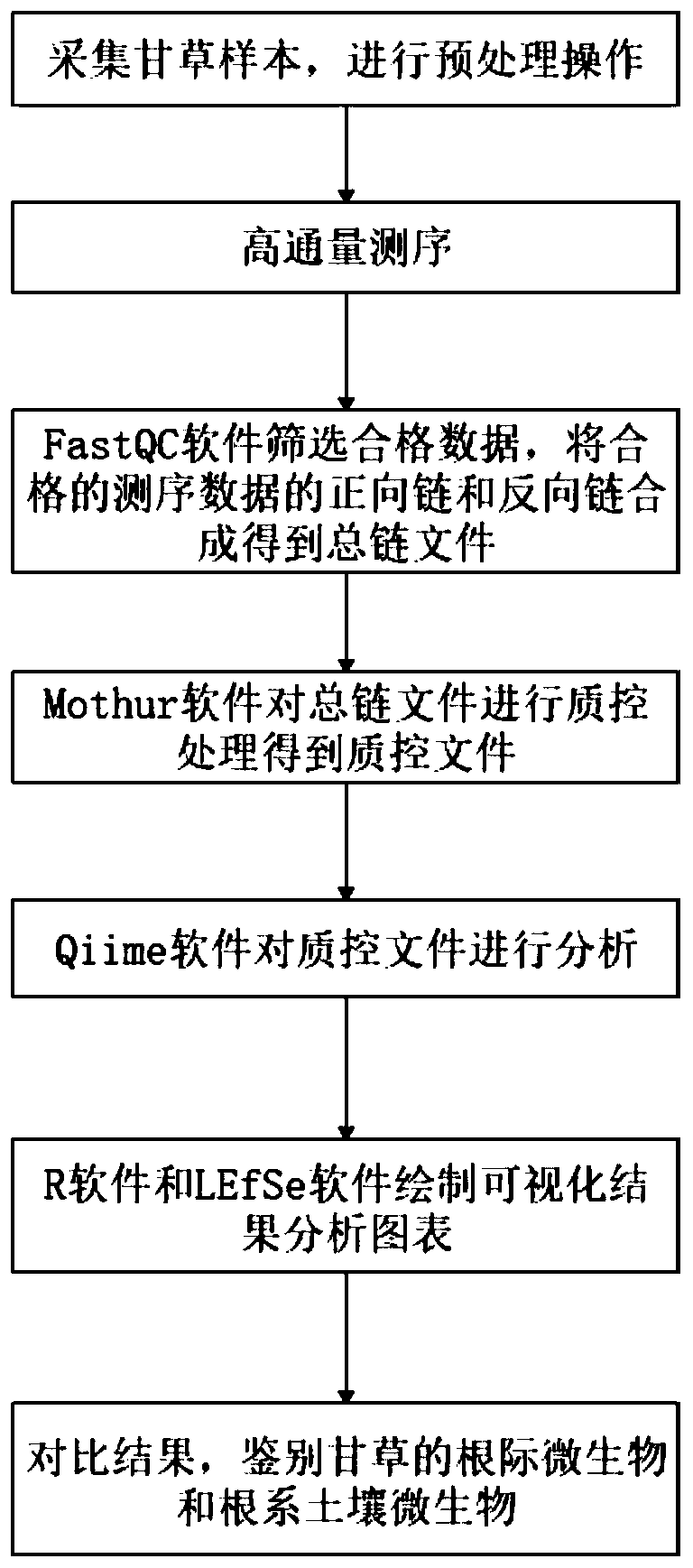
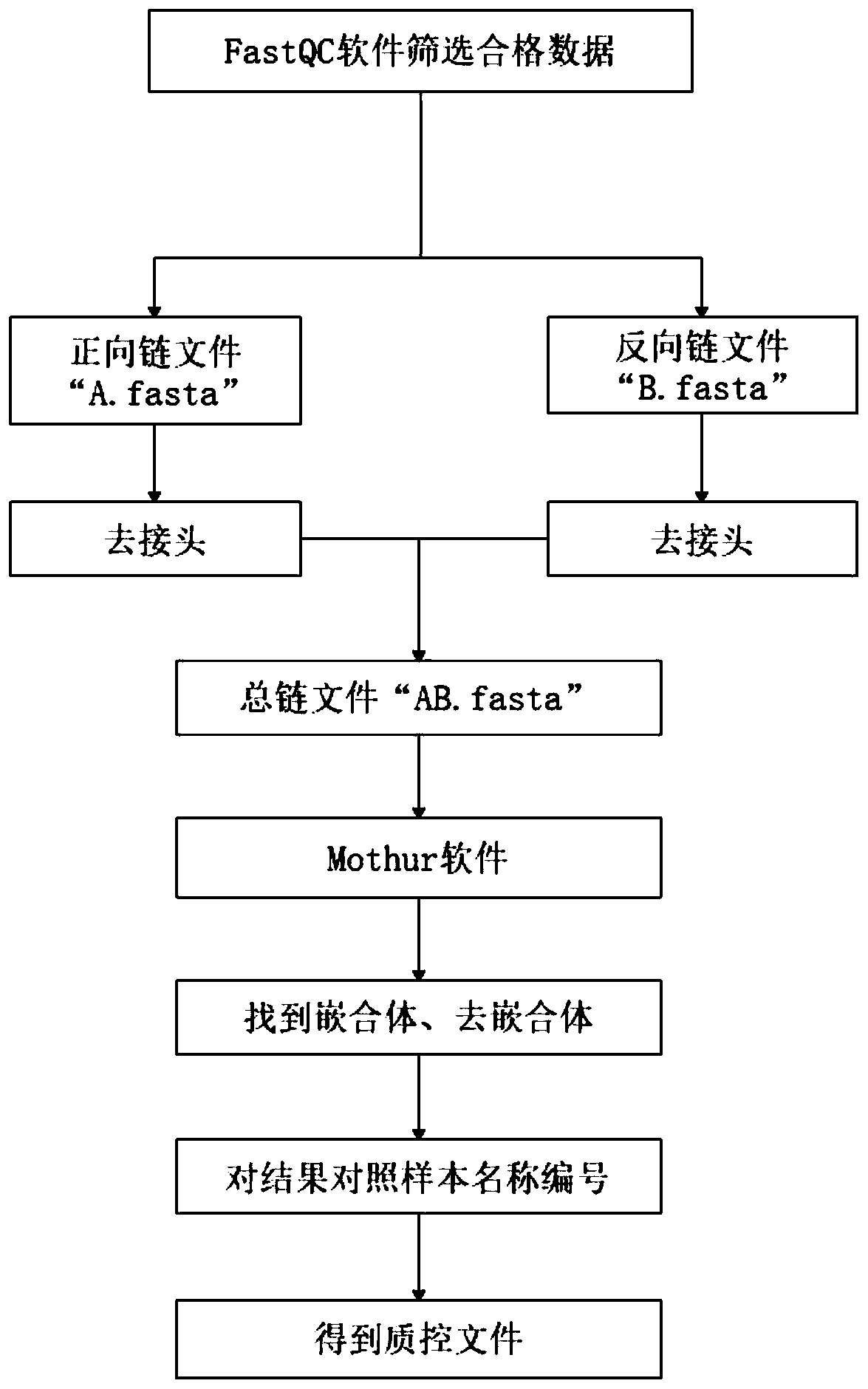
[0047] refer to figure 1 A method for identifying medicinal plant symbiotic microorganisms is applied in identifying licorice rhizosphere microorganisms and root soil microorganisms, which is realized by using a method for identifying medicinal plant symbiotic microorganisms. The method includes the following steps:
[0048] S1, according to different planting methods, collect rhizosphere samples of wild licorice and cultivated licorice, as well as corresponding root soil samples, soak all samples in liquid nitrogen tanks, freeze them, and finally store them in dry ice boxes. The specific operations are as follows:
[0049] The rhizosphere samples of wild licorice and cultivated licorice in Shijiquan Village, Gaoshawo Town, Yanchi County were collected, hereinafter referred to as wild samples and cultivated samples respectively, among which 25 wild samples, 25 cultivated annual samples and 25 cultivated three-year-old samples and corresponding root soil samples, hereinafter re...
Embodiment 2
[0099] The application of a method for identification of medicinal plant symbiotic microorganisms in the identification of licorice planting methods is completed by the method described in Example 1, by Figure 5 ~ Figure 7 It can be seen that the rhizosphere microorganisms and root soil microorganisms of licorice are very different under different planting methods, and the identification of the licorice planting methods to be determined is completed according to the results of comparing the composition of microbial abundance.
Embodiment 3
[0101] The application of a method for identifying medicinal plant symbiotic microorganisms in the analysis of the mechanism of action of traditional Chinese medicine compound prescriptions for treating diseases is realized by the following method, and the method includes the following steps:
[0102] S1, collect the Chinese medicine sample, the subject's genome sample, the stool sample of the subject before taking the Chinese medicine sample, and the stool sample of the subject after taking the Chinese medicine sample, and perform pretreatment operations;
[0103] The specific steps of the pretreatment operation are: collect the Chinese medicine sample, the genome sample of the subject, the stool sample of the subject before taking the Chinese medicine sample, and the stool sample of the subject after taking the Chinese medicine sample, and perform - Freeze below 60°C and store in a dry ice box. The specific operation of the freeze is to immerse all samples in a liquid nitroge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com