Duck circovirus tandem repeated sequence and application thereof
A technology in the sequence and sequence listing, applied in the field of duck circovirus tandem repeats, can solve the problem of few studies on tandem repeats, and achieve the effect of strong promotion value and application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
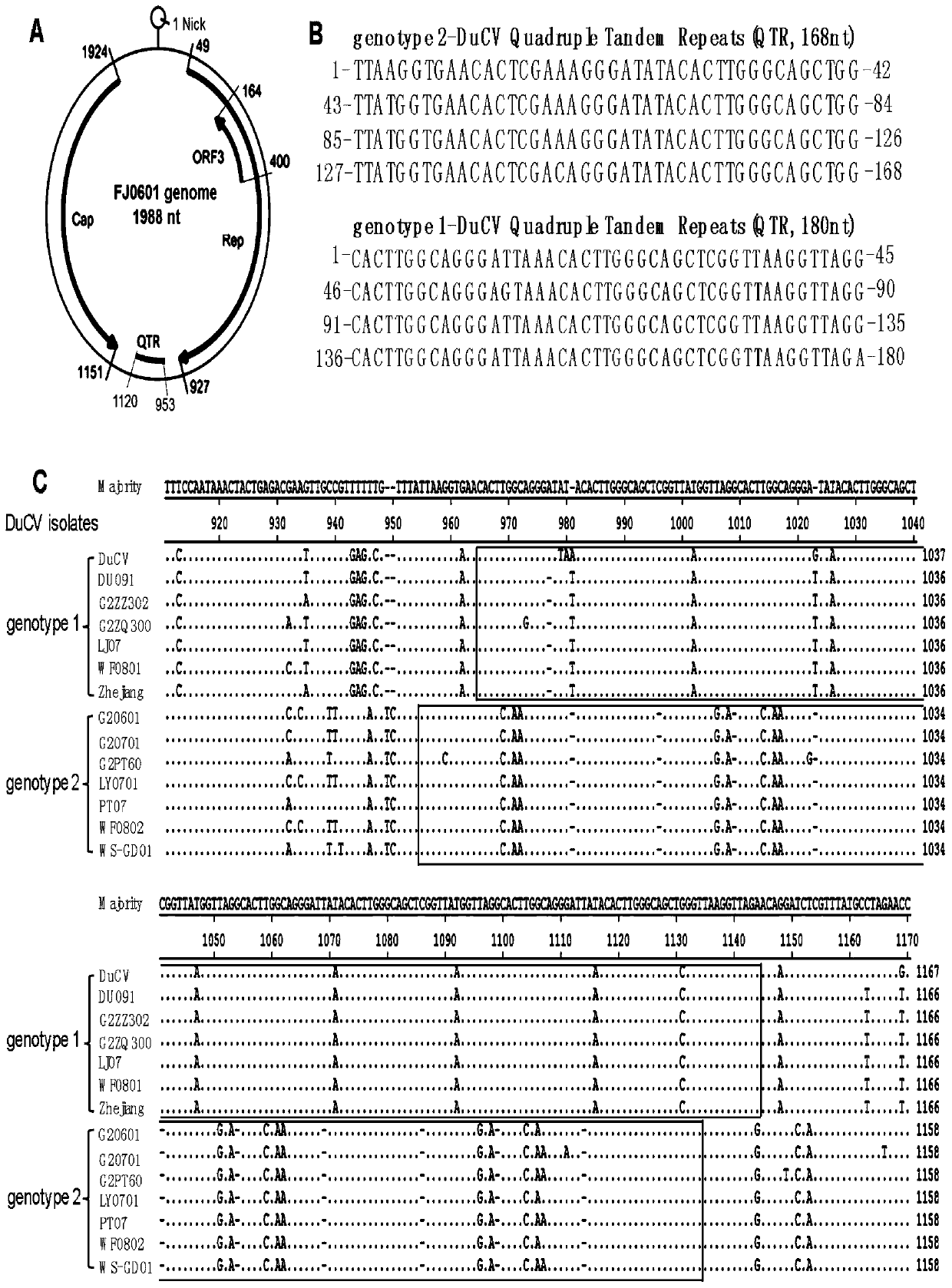
[0073] Example 1 Dottup bioinformatics tool analysis of DuCV repeat sequences
[0074] 1. Using the dottup bioinformatics tool of EMBOSS Program, taking DuCV strain as the representative strain of genotype I and FJ0601 strain as the representative strain of genotype II, analyzed the DuCV genome repeat sequence, and found the middle of gene I and type II DuCV genome There are tandem 4-copy repeat sequences (QTR) in the region, and there are 2 small repeat sequences in each large repeat unit of genotype I QTR;
[0075] 2. In order to analyze whether the original QTR circovirus is conservative, use the dottup tool to select porcine circovirus (PCV1 and PCV2), goose circovirus GoCV, and parrot beak and feather disease virus (PBFDV) with relatively advanced DuCV kinship. Genome analysis did not reveal repetitive sequences.
Embodiment 2
[0076] Example 2 Sequence of tandem repeat QTR, genomic location and comparative analysis of two genotype QTR sequences
[0077] 1. Analysis of the DuCV genome location of QTR found that QTR is located in the middle region of Rep and Cap genes;
[0078] 2. QTR sequence analysis found that the genotype II QTR is 168bp long, including four 42bp tandem repeat units, and the genotype I QTR is 180bp long, including four 45bp tandem repeat units, and each repeat unit contains two 7bp spacer repeat units;
[0079] Genotype I QTR sequence:
[0080] 5'-cacttggcagggattaaacacttgggcagctcggttaaggttaggcacttggcagggagtaaacacttgggcagctcggttaaggttaggcacttggcagggattaaacacttgggcagctcggttaaggttaggcacttggcagggattaaacacttgggcagctcggttaaggttaga-3' (SEQ ID NO. 1)
[0081] Genotype II QTR sequence:
[0082] 5'-ttaaggtgaacactcgaaagggatatacacttgggcagctggttatggtgaacactcgaaagggatatacacttgggcagctggttatggtgaacactcgaaagggatatacacttgggcagctggttatggtgaacactcgacagggatatacacttgggcagctgg-3' (SEQ ID NO. 2)
[00...
Embodiment 3
[0084] Example 3 Analysis of QTR transcription regulation
[0085] 1. Construction of luciferase reporter plasmid: DuCV strain was used as the representative strain of genotype I, FJ0601 strain was used as the representative strain of genotype II, and the genotype I DuCV QTR fragment (G1-QTR) and genotype II were amplified respectively DuCV QTR fragment (G2-QTR);
[0086] 2. Using different restriction sites and seamless cloning technology, insert G1-QTR and G2-QTR forward or reverse into the upstream and downstream regulatory regions of the luciferase gene, respectively
[0087] 3. The constructed luciferase reporter plasmid was transfected into DEF cells, and the pRL-TK plasmid was simultaneously transfected as an internal reference reporter plasmid. After 36 hours of transfection, the cells were collected, and the luciferase activity was detected by a dual-luciferase detection system.
[0088]4. Analysis of transcription regulation function: QTR inserted downstream of poly...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com