A method for constructing an integrated high-efficiency expression of acetaldehyde dehydrogenase Bacillus subtilis
A technology of acetaldehyde dehydrogenase and Bacillus subtilis, which is applied in the construction field of Bacillus subtilis, can solve the problems of high cost and difficulty in large-scale production, and achieve the effect of increasing yield and efficient secretion expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
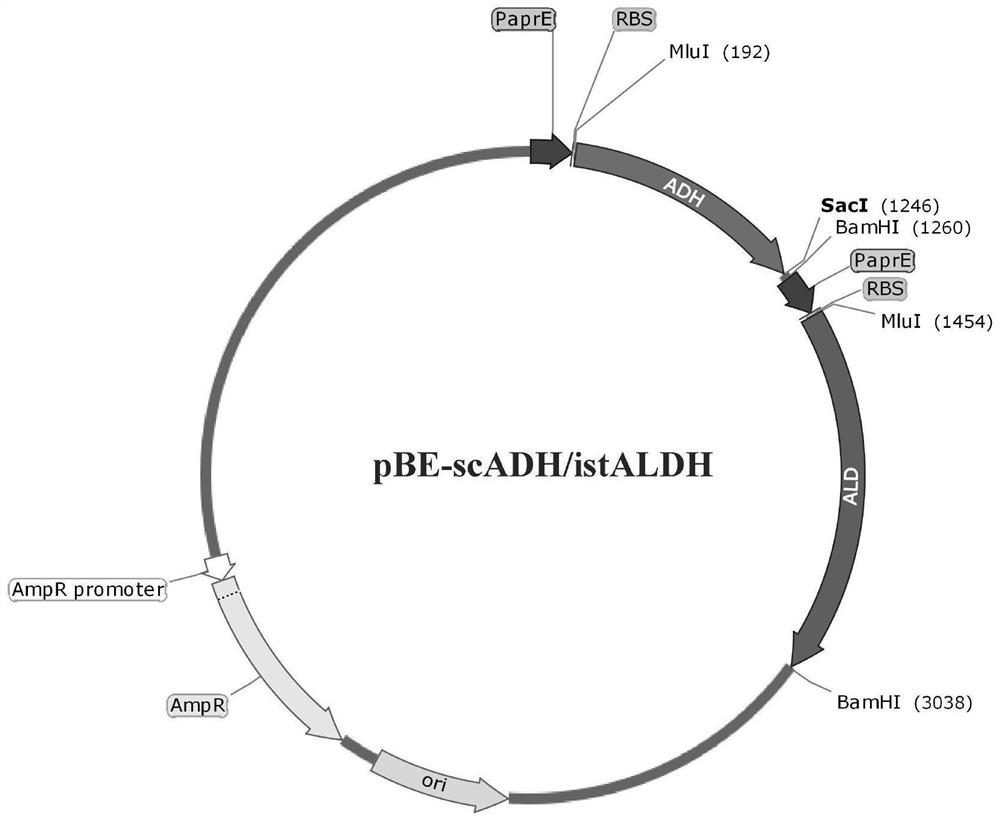
[0033] Example 1: Construction of integrated acetaldehyde dehydrogenase secretion expression vector
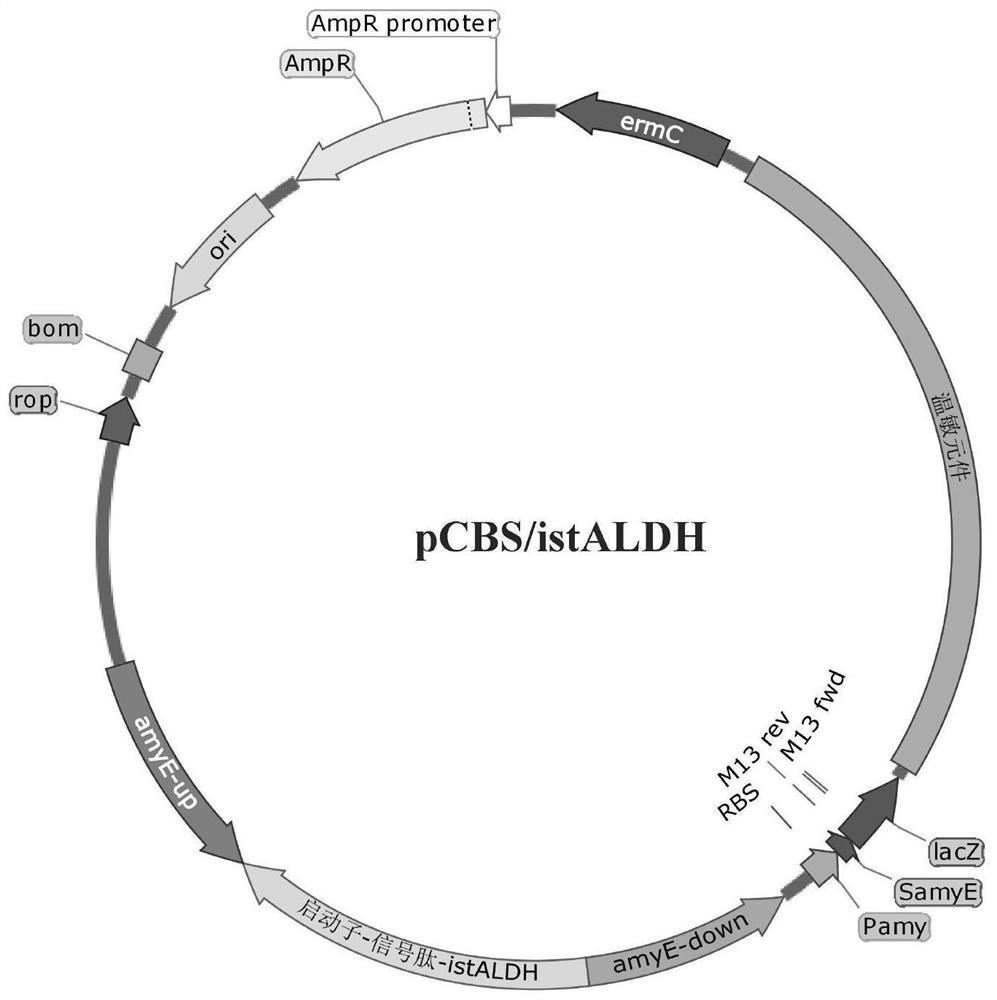
[0034] The integrative plasmid can be any plasmid that can replicate in Bacillus subtilis and Escherichia coli and can be integrated into the chromosome by homologous recombination. The integration site may be the position of amylase (amyE), xylanase (xylA) and any other gene not necessary for the growth of Bacillus subtilis in Bacillus subtilis. The promoter contained in the integrating plasmid can be P amyQ ,P amyE ,P amyL ,P aprE ,P xylA or P glv Any one of or other promoters, and the signal peptide can be any one of SPaprE, SPchiA, SPwapA, SPpbpA or SPyqzG or other signal peptides. In this embodiment, the integration plasmid pCBS is used, the integration site is the amyE gene, and the promoter P amyL -P amyQ -P cry3A Taking the signal peptide SPchiA as an example to describe the construction process of the integrated plasmid, the principles and processes of other ...
Embodiment 2
[0050] Example 2: Construction of Bacillus subtilis with integrated secretory expression of acetaldehyde dehydrogenase
[0051] Transformation of the recombinant plasmid: transform the recombinant plasmid pCBS / istALDH into Bacillus subtilis 168 chemically competent cells, spread the LB resistance plate (containing 5 μg / mL erythromycin), culture overnight at 37°C, and verify the transformants by PCR.
[0052] Screening of recombinant strains: pCBS plasmid is a temperature-sensitive shuttle plasmid for Escherichia coli and Bacillus subtilis, and contains a temperature-sensitive replication origin site for Gram-positive bacteria. At 37°C, autonomous replication can be carried out in the host bacteria, but when the temperature rises to 42°C, autonomous replication cannot be carried out.
[0053] The Bacillus subtilis containing the recombinant plasmid was cultured at 42°C and 180rpm for 24 hours, then spread on an LB plate, cultured at 42°C to obtain a single colony, and induced t...
Embodiment 3
[0058] Example 3: Construction of Bacillus subtilis overexpressing CsaA strain (CsaA gene GenBank: CP041757 REGION: 2078810---2079142, 333bp)
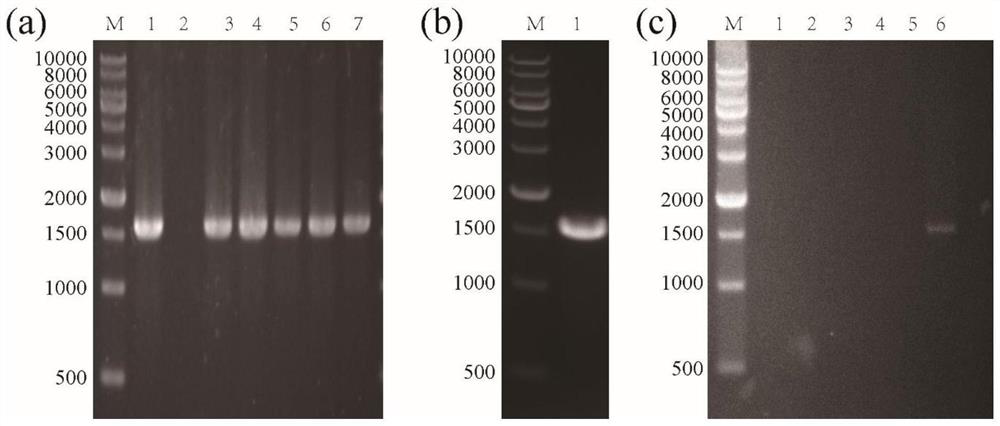
[0059] Using the genomic DNA of Bacillus subtilis 168 as a template, PCR amplification was performed with primers 9 and 10 (underlined restriction sites), and the PCR product (CsaA, 333bp) was recovered. Add A reaction to the recovered product, connect it to pMD19T vector (Takara Company) with T4 ligase, transform Escherichia coli competent cells, and obtain recombinant plasmid pMD19T / csaA (Mlu I-Sal I). Digest pMD19T / csaA (Mlu I-Sal I) and expression plasmid pBE (purchased from Takara, Cat#3380) with restriction endonucleases Mlu I and Sal I, purify the digested product, and use T4 ligase The linearized pBE vector was connected to the csaA fragment, transformed into Escherichia coli competent cells, and the sequenced correct plasmid was named pBE / csaA ( Figure 4 , Figure 6 a&c, the verified fragment is csaA, 333bp).
[0060] Tran...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com