A tumor marker detection kit based on force coding and its application
A kit and coding sequence technology, applied in the field of biochemical analysis, can solve problems such as difficulty in meeting actual detection requirements, low sensitivity, and high technical requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] Embodiment 1: Preparation of aldehydized glass substrate
[0115] The cut glass slices were ultrasonically washed twice with acetone, then ultrasonically washed three times with ultrapure water, ultrasonically washed with 1mol / L NaOH solution for 10min, and then washed twice with ultrapure water and absolute ethanol respectively. Place it in a drying oven at 250°C for 3 hours to remove bacteria on the surface.
[0116] Add the sterilized glass substrate into the newly prepared Piranha solution beaker (98% concentrated H 2 SO 4 : 30%H 2 o 2 =7:3, the volume ratio of the two), put the above-mentioned beaker in a 90°C constant temperature water bath and let it soak for 30min, and the surface of the glass sheet is hydroxylated.
[0117] The glass substrate just hydroxylated above was added into a toluene solution containing 1% APTES, soaked at room temperature for 3 hours, and the glass substrate was aminated. Wash the surface with toluene solution for 3 times, then wa...
Embodiment 2
[0119] The fixation of embodiment 2 nanometer sensor
[0120] Add 20 μL of 3 μM aminated MUC1 force coding sequence and 3 μM aminated HER2 force coding sequence at a volume ratio of 1:1 to the surface of the aldylated glass substrate prepared in Example 1, reduce with sodium borohydride, and incubate overnight at 4°C.
[0121] Mix equal volumes of 3 μM MUC1 aptamer and 3 μM complementary DNA sequence (Com 13), react in a metal bath at 90°C for 5 min, and then allow it to cool down to room temperature naturally to obtain the hybridized MUC1 first hybrid nucleic acid (hereinafter Abbreviated as DNA1). Mix equal volumes of 3 μM HER2 nucleic acid aptamer and 3 μM complementary DNA sequence (Com 16), react in a metal bath at 90°C for 5 min, and then allow it to cool down to room temperature naturally to obtain the hybridized HER2 first hybrid nucleic acid (following Abbreviated as DNA2).
[0122] Take 20 μL of DNA1 and DNA2 with a volume ratio of 1:1 and add them to the sample po...
Embodiment 3
[0123] Embodiment 3 force spectrum measurement
[0124] This embodiment provides a method for force spectrum measurement:
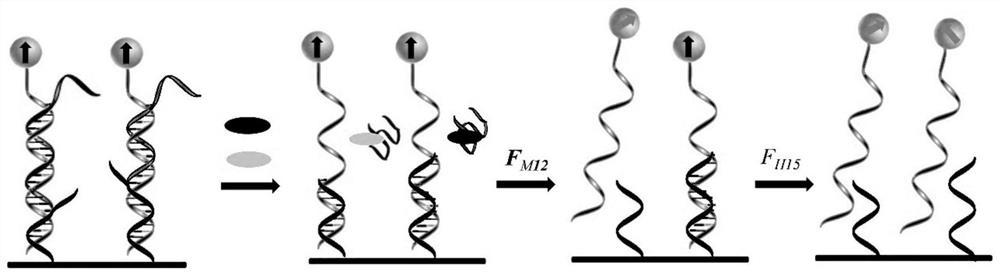
[0125] (1) Fixed MUC1 nanosensor:
[0126] The specific steps are the same as those in Example 2, except that the HER2 nanosensor is not included, and only the MUC1 sensor is fixed on the substrate.
[0127] (2) Measure the force spectrum of MUC1:
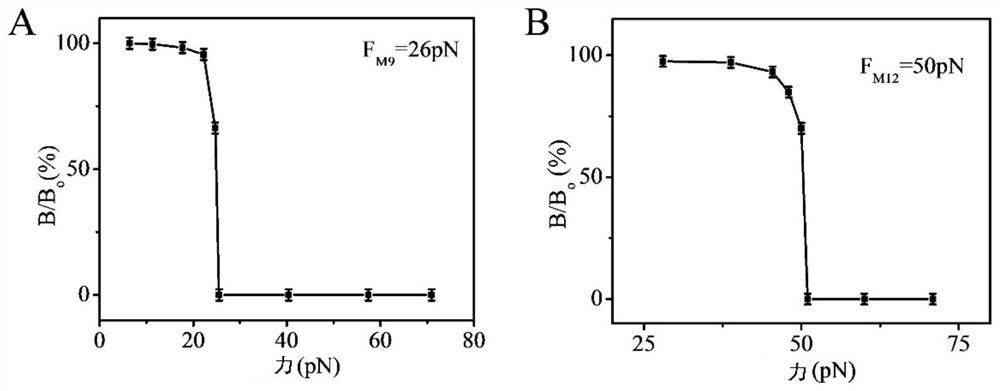
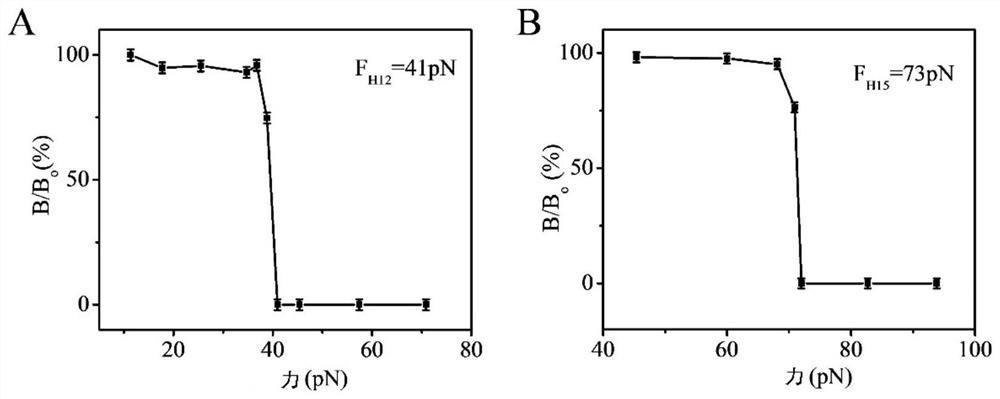
[0128] The glass substrate fixed with the nanosensor obtained in step (1) was centrifuged at 1500 rpm / min for 5 minutes (the direction of the centrifugal force suffered by the nanosensor is directed from the substrate to the nanosensor) to remove physical adsorption. The magnetic signal is then detected on an ultra-low field atomic magnetometer. Gradually increase the magnitude of the centrifugal force, and the centrifugation time is still 5 minutes each time, and record the magnetic signal intensity corresponding to each centrifugal force. Due to the non-covalent bond between double-stranded DNA, under th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com