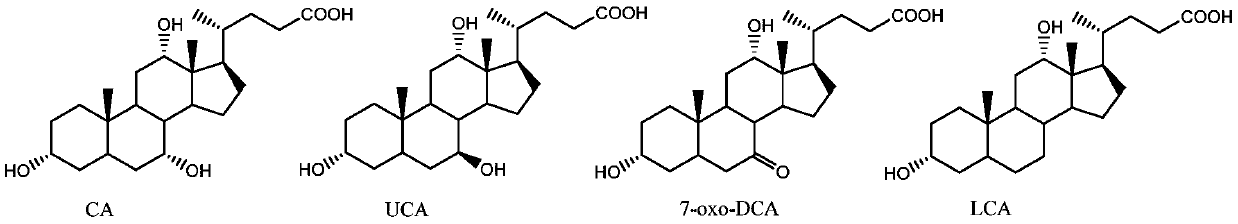
Hydroxysteroid dehydrogenase and application thereof in synthesis of ursodeoxycholic acid precursor
A technology of hydroxysteroids and dehydrogenases, applied in the direction of application, microbial-based methods, enzymes, etc., can solve the problem of high cost of coenzyme application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Gene cloning and transformation expression of hydroxysteroid dehydrogenase Rr12α-HSDH
[0058] The complete genome of Rhodococcus ruber with the preservation number of CGMCC No.1.10360 was obtained by the high-salt method. Inoculate Rhodococcus ruber with the preservation number of CGMCC No.1.10360 into LB medium, incubate at 37°C for 24 hours, take 100 mL of the bacterial solution, centrifuge at 8,000 rpm for 10 minutes, collect the bacteria, wash with 20 mL of saline, and repeat Twice, then resuspend the bacteria in 20 mL of saline. Add 1mL of lysozyme solution (50mg / mL) to the suspension bacteria, and keep the temperature in a 37℃ water bath for 1h; add 1.6mL of sodium lauryl sulfonate solution (10%, w / v), 160μL of proteinase K (20mg / mL) , Keep the temperature in 55℃ water bath until the suspension becomes clear. Add 1 / 3 volume of saturated NaCl solution, shake and mix until the solution becomes turbid, then centrifuge at high speed for 10 minutes to discard...
Embodiment 2
[0060] Example 2 Random mutation of hydroxysteroid dehydrogenase Rr12α-HSDH
[0061] Based on the gene mining of the enzyme in Example 1, error-prone PCR technology was used for random mutation to further improve the activity of the enzyme.
[0062] According to the open reading frame of Rr12α-HSDH, the upstream and downstream primers are designed as follows:
[0063] Upstream primer, as shown in SEQ ID No. 3:
[0064] 5’-CCG GAATTC ATGAAACTGCGCGGGAAGA-3’
[0065] Downstream primer, as shown in SEQ ID No. 4:
[0066] 5’-CCC AAGCTT TCACCGCAGCTTGATGCTGC-3’
[0067] The sequence underlined in the upstream primer is the restriction site of EcoRI, and the sequence underlined in the downstream primer is the restriction site of Hind III.
[0068] Using pET28a-12α-HSDH as a template, error-prone PCR was performed with rTaq DNA polymerase to construct a random mutation library.
[0069] The PCR system (50μL) contains the following components:
[0070] Taq DNA polymerase 0.5μl, 10×PCR buffer(Mg 2+ P...
Embodiment 3
[0091] Example 3 Hydroxysteroid dehydrogenase Rr12α-HSDH M8 Gene cloning
[0092] On the basis of the gene cloning described in Example 1, according to the hydroxysteroid dehydrogenase Rr12α-HSDH M8 Design the upstream and downstream primers, and use the nucleic acid sequence numbered as SEQ ID No. 6 as the template for PCR amplification.
[0093] Upstream primer, as shown in SEQ ID No. 3:
[0094] 5’-CCG GAATTC ATGAAACTGCGCGGGAAGA-3’
[0095] Downstream primer, as shown in SEQ ID No. 4:
[0096] 5’-CCC AAGCTT TCACCGCAGCTTGATGCTGC-3’
[0097] The underlined sequence of the upstream primer is the restriction site of EcoRI, and the underlined sequence of the downstream primer is the restriction site of Hind III.
[0098] The PCR system is: 2×Taq PCR MasterMix 25μL, upstream primer and downstream primer (10ng / μL) each 2.5μL, 1μL plasmid containing the nucleic acid sequence numbered SEQ ID No.5 gene (150ng / μL), and 19μL ddH 2 O. The PCR amplification program is: pre-denaturation at 95°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com