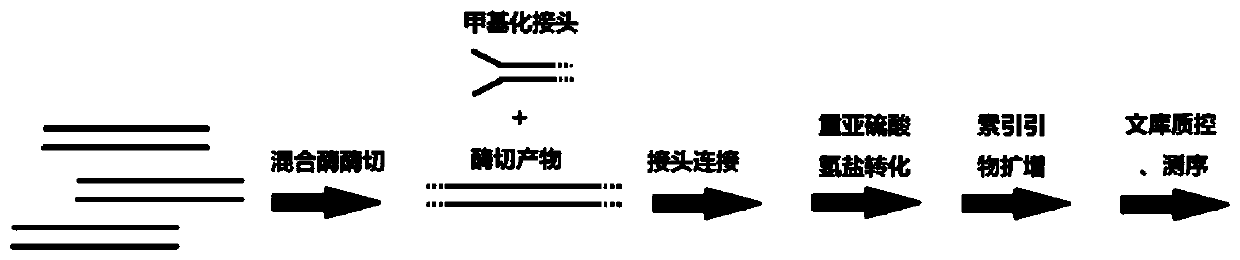
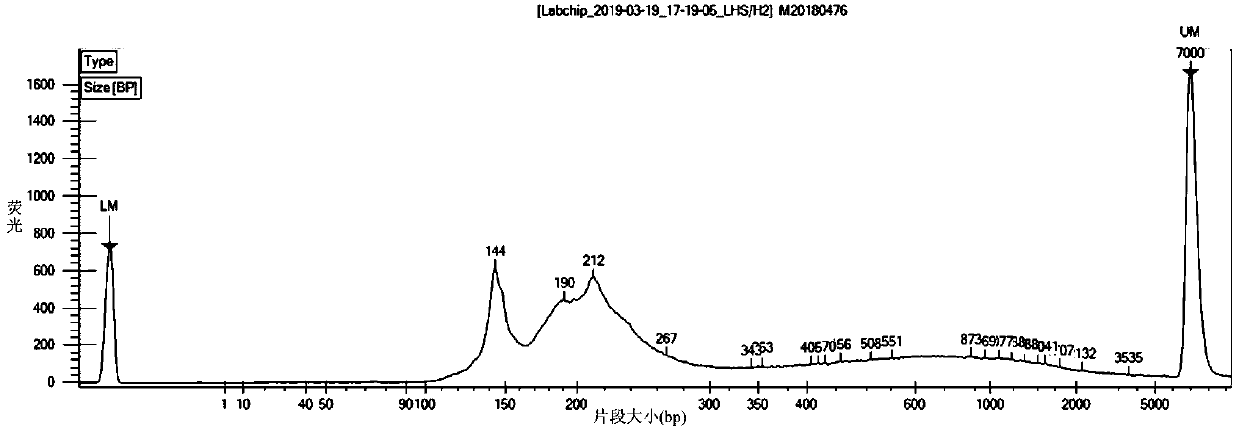
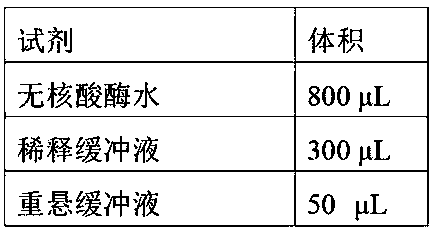
DNA methylation marker screening kit and method
A methylation and kit technology, applied in the field of molecular biology, can solve the problems of low enrichment of CpG sites, large initial amount of DNA library construction, and low site reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0125] one. Magnetic bead purification of extracted DNA
[0126] 1. Take out the AMPure magnetic beads half an hour in advance and place them at room temperature. After vortexing the magnetic beads, add 50 microliters of magnetic beads to 50 microliters of DNA solution, and mix by pipetting. Let stand at room temperature for 5 minutes.
[0127] 2. Transfer to a magnetic stand until the liquid is completely clear.
[0128] 3. Keep on the magnetic stand, discard the supernatant, and wash the magnetic beads with 150 μl of freshly prepared 80% ethanol.
[0129] 4. Repeat step 3.
[0130] 5. Remove the ethanol completely and dry with the lid open for 5 minutes.
[0131] 6. Remove the sample from the magnetic stand, add 32 microliters of nuclease-free water, fully resuspend the magnetic beads with a pipette gun, and place at room temperature for 5 minutes.
[0132] 7. Transfer samples to a magnetic stand until completely clear.
[0133] 8. Transfer 30 µl of eluate to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com