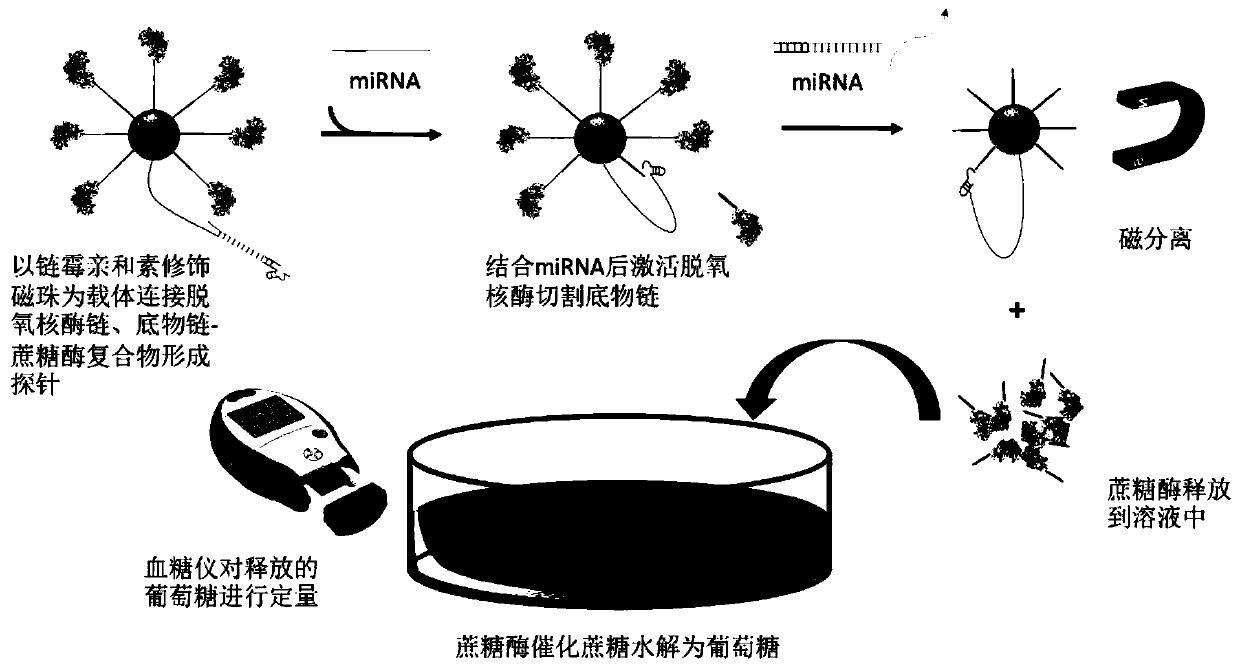
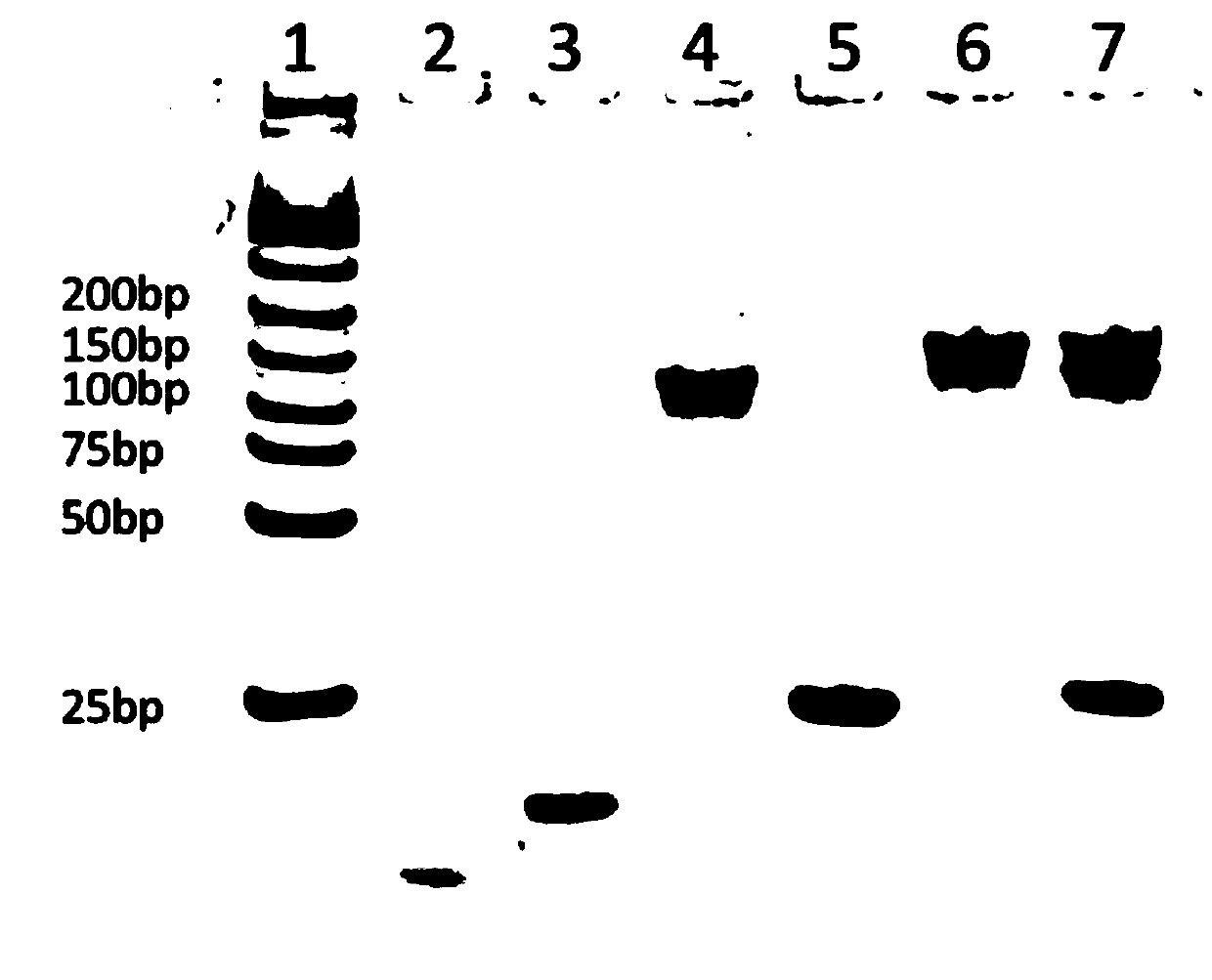
Method for detecting miRNA-21 by blood glucose meter based on dnazyme and sucrase
A technology of miRNA-21 and deoxyribozyme, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of miRNA high NGS error rate, limited clinical application, lengthy time, etc., to improve detection sensitivity, Highly modifiable, low-cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Feasibility verification of the detection method of the present invention, evaluation of the competition and hybridization feasibility of the system design sequence in a buffer solution
[0040] 1. Preparation of substrate strand and sucrase-linked complex
[0041] Prepare 30 μL of substrate chain with a concentration of 1 mM in DEPC water, add 2 μL of sodium phosphate buffer (1M) at pH 5.5 and 2 μL of tris(2-carboxyethyl)phosphine (TCEP) at a concentration of 30 mM dissolved in Millipore water, and The mixture was allowed to stand at room temperature for 1 hour and then purified by ultrafiltration 8 times using Amicon-10K ultrafiltration tubes.
[0042] Sucrase-linked substrate chains: Prepare 400 μL of 20 mg / mL sucrase solution with buffer A, mix with 1 mg sulfo-SMCC; vortex for 5 minutes, place on a shaker at room temperature for 1 hour, and centrifuge at 1000r for 5 minutes Excess sulfo-SMCC was removed and the solution was purified by ultrafiltration 8 ...
Embodiment 2
[0053] Embodiment 2: investigate the sensitivity of the present invention to miRNA-21
[0054] 1. The instruments and reagents required for the experiment are the same as those in Implementation 1;
[0055] 2. Investigate the sensitivity of the deoxyribozyme and sucrase dual-enzyme amplification system to measure the miRNA-21 standard solution, including the following steps:
[0056] (1) Preparation of samples: prepare standard solutions of miRNA-21 at concentrations of 100fM-1000fM with DEPC water, so that the concentrations are 100fM, 200fM, 400fM, 600fM, 800fM, and 1000fM;
[0057] (2) Start the reaction: Add 10 μL miRNA-21 standard solution and 60 μL probe dissolved in buffer C to each tube, and then add Mn dissolved in DEPC water at a concentration of 0.01M 2+ solution; incubate at room temperature for 40min;
[0058] (3) Stop the reaction: Add 5 μL of 50 mM EDTA to stop the reaction;
[0059] (4) Magnetic separation: Adsorb the magnetic beads with a magnet, transfer t...
Embodiment 3
[0064] Embodiment 3: investigate the selectivity of the present invention to miRNA-21
[0065] 1. The instruments, reagents and probe synthesis methods required for the experiment are consistent with those in Implementation 1;
[0066] 2. The sequences required for this experiment, in addition to the sequences applying for rights protection, also need to use the following sequences:
[0067] name oligonucleotide sequence has-mir-200c 5'-cgtcttacccagcagtgtttgg-3' hsa-mir-423 5'-tgaggggcagagagcgagacttt-3' hsa-miR-4640 5'-tgggccagggagcagctggtggg-3' has-miR-21 5'-uagcuuaucagacugauguuga-3' Mis-1 5'-uaccuuaucaacacugaucuuga-3' Mis-2 5'-uagcuuaucagacagauguuga-3' Mis-3 5'-uagcuuaacagacugauguuga-3'
[0068] 3. Deoxyribozyme and sucrase-based dual-enzyme amplification system measures the selectivity of miRNA-21 and mismatched sequences, comprising the following steps:
[0069] (1) Preparation of samples: Prepare hsa-miR-20...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com