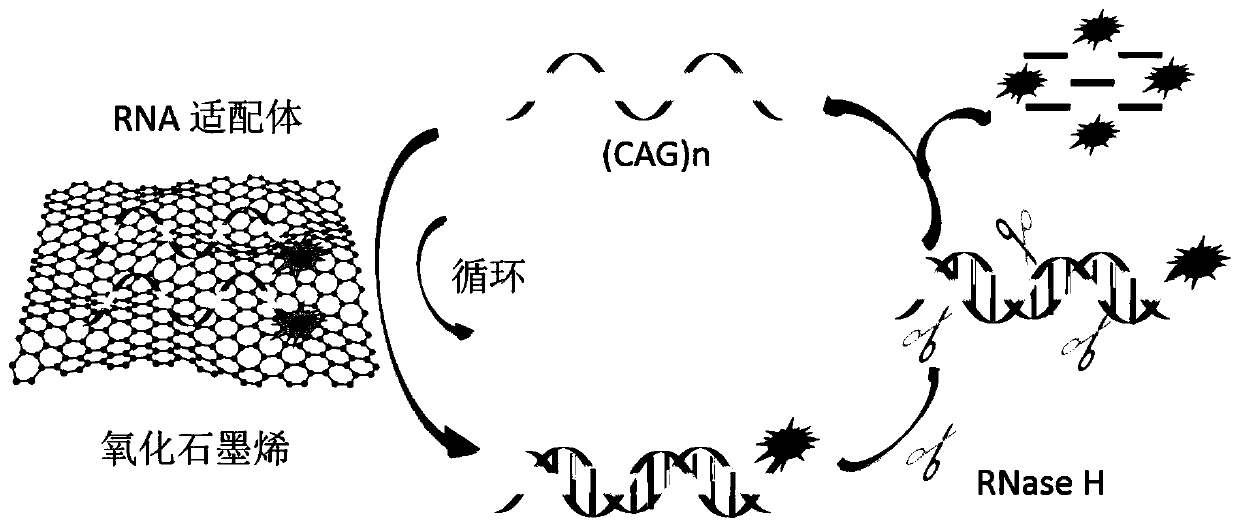
Method for detecting (CAG)n repeated sequence by utilizing RNase H
A detection method and nucleotide sequence technology, applied in biochemical equipment and methods, biological testing, measuring devices, etc., can solve the problems of target DNA that cannot be reused, less fluorescent groups are difficult to detect, and detection accuracy is poor , to achieve the effect of enhanced detection sensitivity, high sensitivity and rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Feasibility analysis of working principle:
[0046] (1) Dilute graphene oxide to 100 μg / ml with DEPC water, mix well, and set aside.
[0047] (2) Dissolve the fluorescently labeled aptamer R1 with DEPC water (sequence is 5'-FAM-GUCGUC GUC GUC GUCGUC GUC GUC GUC GUC-3') to prepare 20×10 -6 mol / L solution, mix well and set aside.
[0048] (3) Dissolve (CAG)n (sequence is 5'-CAGCAG CAG CAG CAG CAG CAG CAG CAG CAG CAG-3') with DEPC water to prepare 100×10 -6 mol / L solution, mix well and set aside.
[0049] (4) Dilute the fluorescently labeled aptamer R1 solution prepared in step (2) with DEPC water to 200×10 -9 mol / L, mix well, and become tube ①.
[0050] (5) Add the graphene oxide solution prepared in step (1) with a final concentration of 24 μg / ml into tube ①, mix well, and incubate at room temperature for 10 min to form tube ②.
[0051] (6) Add final concentration of 25×10 to tube ② -9 mol / L of (CAG)n prepared in step (3), mix well, incubate at room temperature for...
Embodiment 2
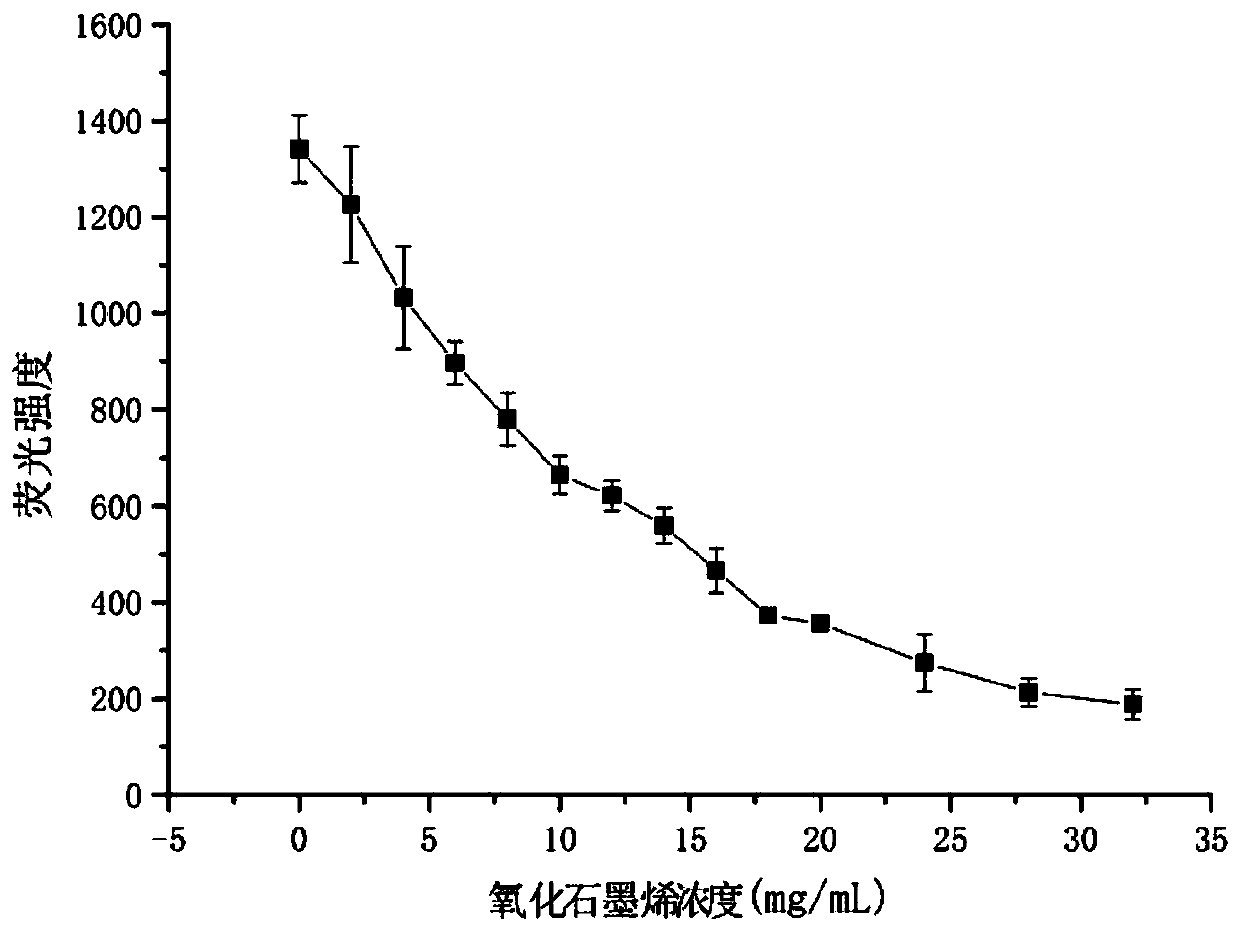
[0056] The ability of graphene oxide concentration to quench aptamer RNA fluorescence:
[0057] 1. Dilute graphene oxide to 100μg / ml with DEPC water and mix well.
[0058] 2. Dissolve the fluorescently labeled aptamer R1 (sequence is 5'-FAM-GUCGUC GUC GUC GUC GUCGUC GUC GUC GUC GUC-3') with DEPC water to prepare 20×10 -6 mol / L solution and mix well.
[0059] 3. Add different final concentrations of graphene oxide solution (0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 24, 28, 32 μg / ml) to Tris-HCl buffer solution respectively 20×10 in -6 mol / L FAM-R1, mix well, and incubate at room temperature for 30 min.
[0060] 4. Use a Hitachi F-7000 spectrofluorometer. The excitation wavelength was 480 nm, the excitation slit was 5 nm, the emission slit was 2.5 nm, the scanning speed was 1200 nm / min, and the fluorescence intensity at 520 nm was detected. The results showed that with the increase of graphene oxide concentration, the fluorescence intensity decreased continuously. After 24 μg / ...
Embodiment 3
[0063] To investigate the effect of aptamer fluorescence quenching time, the specific steps are as follows:
[0064] 1. Dilute graphene oxide to 100μg / ml with DEPC water and mix well.
[0065] 2. Dissolve the fluorescently labeled aptamer R1 (sequence is 5'-FAM-GUCGUC GUC GUC GUC GUCGUC GUC GUC GUC GUC-3') with DEPC water to prepare 20×10 -6 mol / L solution and mix well.
[0066] 3. Dissolve (CAG)n (sequence is 5'-CAGCAG CAG CAG CAG CAG CAG CAG CAG CAG CAG-3') with DEPC water to prepare 100×10 -6 mol / L solution and mix well.
[0067] 4. In containing 200×10 -9 Add 24 μg / ml graphene oxide solution to the Tris-HCl buffer solution of mol / L FAM-R1, mix well, and incubate at room temperature for 0, 2, 5, 8, 10, 12, 15, 20, 25, and 30 min respectively.
[0068] 5. Use a Hitachi F-7000 spectrofluorometer. The excitation wavelength was 480 nm, the excitation slit was 5 nm, the emission slit was 2.5 nm, the scanning speed was 1200 nm / min, and the fluorescence intensity at 520 nm wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com