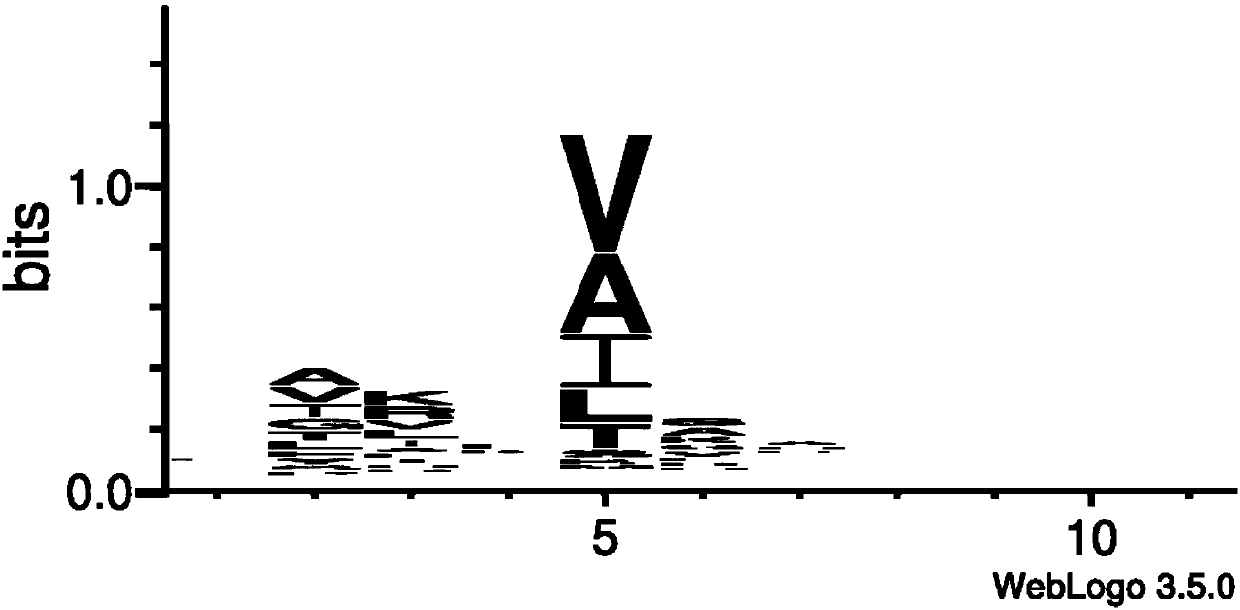
Protein analysis method based on hydrophobic amino acid specific protease
A technology of hydrophobic amino acid and analysis method, which is applied in the field of protein analysis based on hydrophobic amino acid-specific protease, and achieves the effect of improving the accuracy and application range, and increasing the number of identifications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Short-chain hydrophobic amino acid-specific protease for the analysis of myoglobin
[0017] (1) Take 0.2mg of Myoglobin and dissolve it in 200μL of 50mM Tris-HCl, 8M Urea, 150mMNaCl, 20mM CaCl 2 In the solution, the pH is 5.0, so that the concentration of Myoglobin in the solution is 1 mg / mL;
[0018] (2) Add the Myoglobin solution system in the above step (1) to a 30K ultrafiltration tube, and centrifuge at 10000g for 30min;
[0019] (3) Add 100 μL of 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl to the ultrafiltration tube of the above step (2) 2 solution, the pH is 5.0, the Urea concentration is lower than 2M, the ratio of protein to enzyme is 20:1, add 10 μg of the protease, and incubate overnight at 20°C;
[0020] (4) Centrifuge the ultrafiltration tube with the Myoglobin solution after enzymolysis in the above step (3) at 10000 g for 30 min, and collect about 100 μL of proteolysis solution;
[0021] (5) Add 100 μL of 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl to the ult...
Embodiment 2
[0025] Hydrolysis of bovine serum albumin (BSA) with short-chain hydrophobic amino acid specific protease and detection
[0026] (1) Take 0.2mg of BSA, dissolve in 200μL of 200mM Tris-HCl, 8M Urea, 200mMNaCl, 1mMCaCl 2 In the solution, the pH is 9.0, so that the BSA concentration in the solution is 1mg / mL;
[0027] (2) Add 500mM DTT 16μL to the BSA solution obtained in the above step (1) to make the final concentration of DTT 40mM, incubate at 100°C for 5min, then add 500mM IAA32μL, the final concentration is 80mM, and incubate at 25°C in the dark for 40min , to open the disulfide bond;
[0028] (3) Add the BSA solution system processed in the above step (2) into a 30K ultrafiltration tube, and centrifuge at 10000g for 30min;
[0029] (4) Add 100 μL of 200 mM Tris-HCl, 200 mM NaCl, 1 mM CaCl to the ultrafiltration tube of the above step (3) 2 solution, the pH is 9.0, the concentration of Urea is lower than 0.5M, the ratio of protein to enzyme is 50:1, add 4 μg of the protea...
Embodiment 3
[0035] The proteolytic enzyme Scanase digests the whole protein of HeLa cells and detects:
[0036] (1) HeLa cells were lysed to extract 0.2 mg of whole protein, with a volume of 200 μL, added to a 30K ultrafiltration tube, and centrifuged at 10,000 g for 30 min;
[0037] (2) Add 200μL of 50mM Tris-HCl, 8M Urea, 150mMNaCl, 20mM CaCl to the ultrafiltration tube of the above step (1) 2 Solution, pH 8.0, centrifuged at 10000g for 30min;
[0038] (3) repeat above-mentioned step (2) twice again;
[0039] (4) Add 200 μL of 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl to the ultrafiltration tube of the above step (3) 2 Solution, pH 8.0, centrifuged at 10000g for 30min;
[0040] (5) repeat above-mentioned step (4) twice again;
[0041] (6) Add 100 μL of 50 mM Tris-HCl, 150 mM NaCl, 20 mM CaCl to the ultrafiltration tube of the above step (5) 2 solution, pH 8.0;
[0042] (7) Add 8 μL of 500 mM DTT to the whole protein solution of HeLa cells obtained in the above step (6) to make the f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com