Method for detecting one or more target nucleic acid sequences to be tested by single tube and kit thereof
A nucleic acid sequence and kit technology, applied in the field of molecular biology, can solve problems such as inability to distinguish between specific amplification and non-specific amplification, inability to perform multiple amplifications, unfavorable product promotion, etc., to achieve objective result judgment and high detection results. Effective and reliable, the effect of increased sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
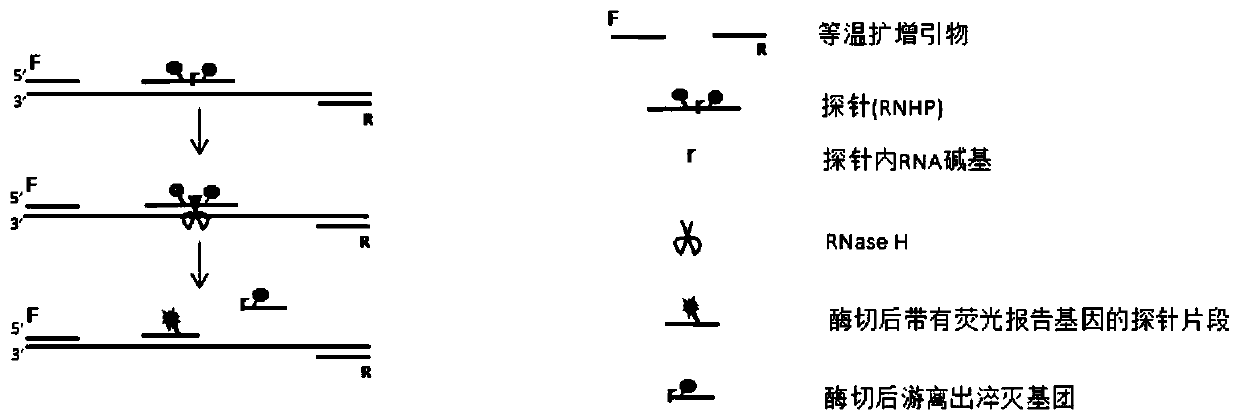
[0035] Embodiment 1 The basic principle of the method of the present invention
[0036] Such as figure 1 As shown, the principle of the method for detecting one or more target nucleic acid sequences to be tested using the fluorescent signal generated by the combination of fluorescent probe (RNHP) and isothermal amplification method in the present invention is as follows figure 1 As shown, the method steps are:
[0037] 1) Design an RNHP for each target nucleic acid sequence to be tested. According to the different isothermal amplification methods used, design specific isothermal amplification primers for each target nucleic acid sequence to be tested, and add the primers and RNHP to the used isothermal amplification Method of the reaction system.
[0038] 2) In the presence of target nucleic acid, as the amplification reaction proceeds, a large amount of target nucleic acid is expanded, and RNHP will bind to the target sequence to form a DNA-RNA hybrid chain. RNaseH can specifically...
Embodiment 2
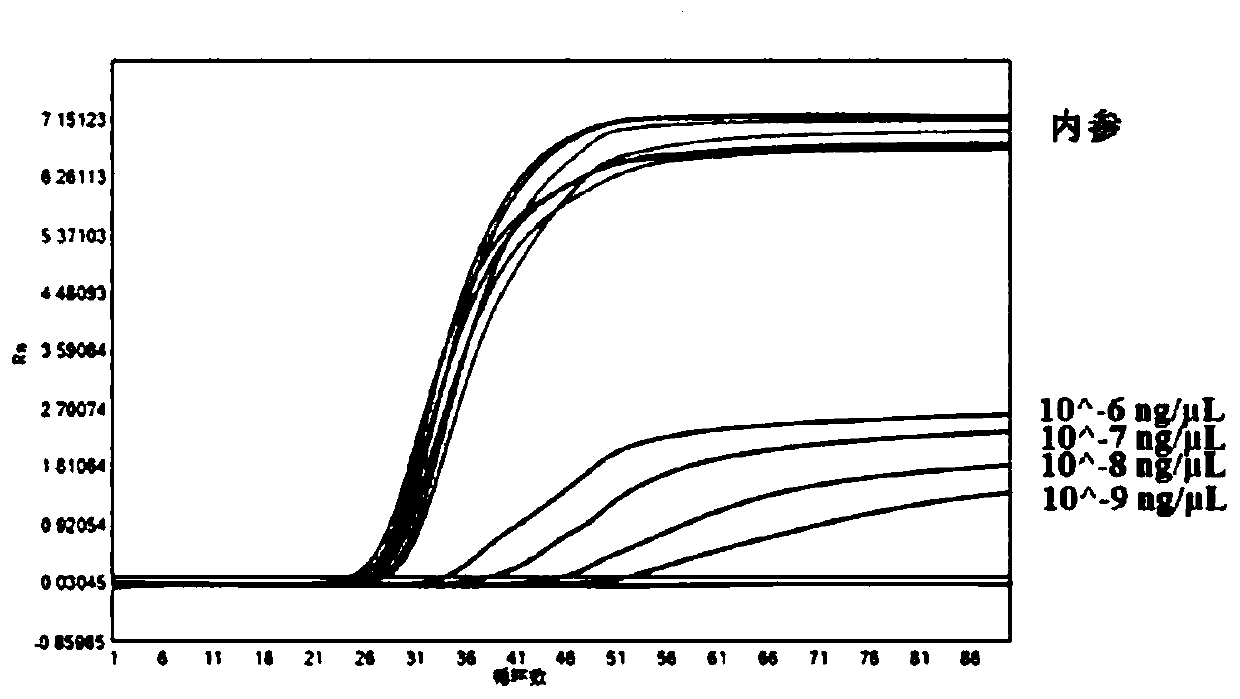
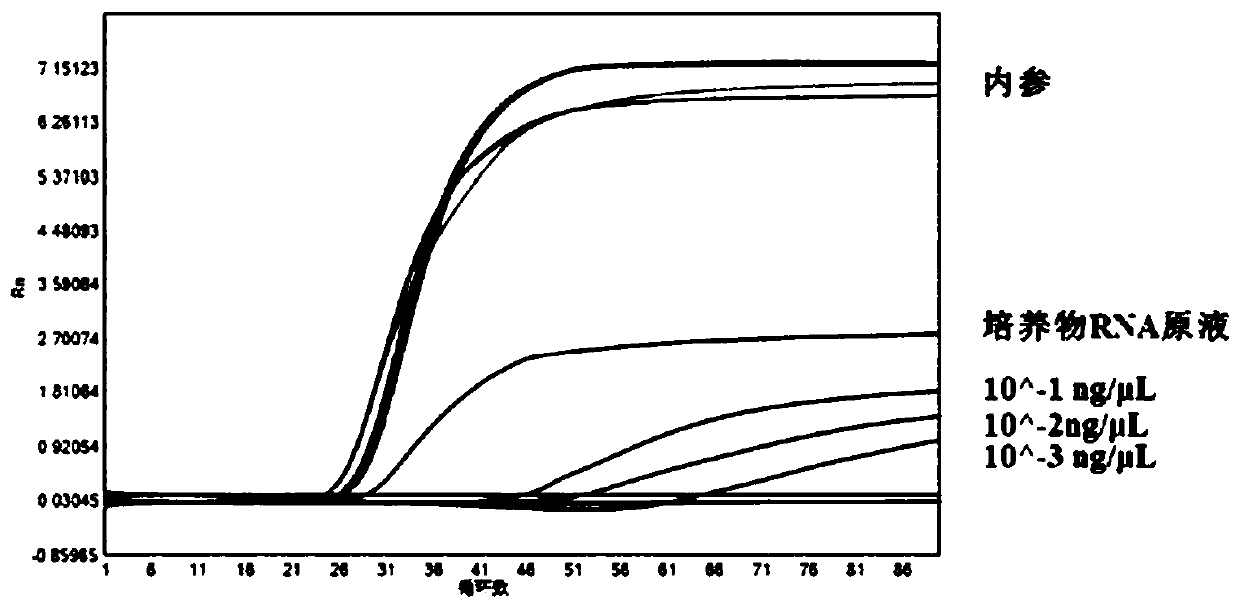
[0039] Example 2 Results of the detection of Zika virus cRNA gradient dilutions and 1 strain of Zika culture RNA gradient dilutions by the real-time LAMP kit of the present invention
[0040] 1. Design of detection primers
[0041] The detection primers are designed using the relevant nucleic acid sequence of Zika virus published on NCBI and the sequence of the internal reference ACTB gene. The detection primer sequences used in the examples are shown in the following table:
[0042]
[0043] Note: In SEQ ID NO:7 and SEQ ID NO:14 in the above table, underlined bases indicate bases that are labeled with fluorescent groups, lowercase bases indicate RNA bases, and the quencher is labeled 3' On the terminal base.
[0044] 2. Preparation of positive quality control products
[0045] Download the human ACTB gene sequence fragment and the Zika virus NS5 gene sequence fragment from NCBI, respectively, design the primers for the amplification of these two fragments: ACTB-F, ACTB-R; ZK-NS5-F, ZK...
Embodiment 3
[0055] Example 3 Amplification of single-plex real-time LAMP is performed when RNHP is designed at different positions in the LAMP amplification region
[0056] 1. Design of detection primers and probes
[0057] The detection primers are designed using the relevant nucleic acid sequences of Bunya virus published on NCBI. The primers and probe sequences used in the examples are shown in the following table:
[0058]
[0059] Note: In SEQ ID NO: 21-25 in the above table, underlined bases indicate bases labeled with fluorescent groups, lowercase bases indicate RNA bases, and the quencher group is labeled on the 3'end base .
[0060] 2. Preparation of positive quality control products
[0061] Download a fragment of the S segment gene sequence of Bunia virus from NCBI, and design the primers for amplification of this fragment to be Bun-S-F (SEQ ID NO: 30, ATTGCTGCTTACAGGTTTCT) and Bun-S-R (SEQ NO: 31, AGGAAAGACGCAGAGGAGTG). Insert the amplified target fragment of Bunia virus into the pMD1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com